Patient-specific Boolean models of signalling networks guide personalised treatments
- ArnauMontagud[email protected]1234
- JonasBéal123
- LuisTobalina5
- PaulineTraynard123
- VigneshwariSubramanian5
- BenceSzalai56
- RóbertAlföldi7
- LászlóPuskás7
- AlfonsoValencia48
- EmmanuelBarillot123
- JulioSaez-Rodriguez59
- LaurenceCalzone[email protected]123
- Research Article
- Computational and Systems Biology
- personalised medicine
- logical modelling
- prostate cancer
- personalised drug
- simulations
- drug combinations
- Human
- publisher-id72626
- doi10.7554/eLife.72626
- elocation-ide72626
Abstract
Prostate cancer is the second most occurring cancer in men worldwide. To better understand the mechanisms of tumorigenesis and possible treatment responses, we developed a mathematical model of prostate cancer which considers the major signalling pathways known to be deregulated. We personalised this Boolean model to molecular data to reflect the heterogeneity and specific response to perturbations of cancer patients. A total of 488 prostate samples were used to build patient-specific models and compared to available clinical data. Additionally, eight prostate cell line-specific models were built to validate our approach with dose-response data of several drugs. The effects of single and combined drugs were tested in these models under different growth conditions. We identified 15 actionable points of interventions in one cell line-specific model whose inactivation hinders tumorigenesis. To validate these results, we tested nine small molecule inhibitors of five of those putative targets and found a dose-dependent effect on four of them, notably those targeting HSP90 and PI3K. These results highlight the predictive power of our personalised Boolean models and illustrate how they can be used for precision oncology.
Introduction
Like most cancers, prostate cancer arises from mutations in single somatic cells that induce deregulations in processes such as proliferation, invasion of adjacent tissues and metastasis. Not all prostate patients respond to the treatments in the same way, depending on the stage and type of their tumour 19Chen and Zhou2016 and differences in their genetic and epigenetic profiles 108Toth et al.2019116Yang et al.2018. The high heterogeneity of these profiles can be explained by a large number of interacting proteins and the complex cross-talks between the cell signalling pathways that can be altered in cancer cells. Because of this complexity, understanding the process of tumorigenesis and tumour growth would benefit from a systemic and dynamical description of the disease. At the molecular level, this can be tackled by a simplified mechanistic cell-wide model of protein interactions of the underlying pathways, dependent on external environmental signals.
Although continuous mathematical modelling has been widely used to study cellular biochemistry dynamics (e.g. ordinary differential equations) 44Goldbeter200258Kholodenko et al.199569Le Novère201599Sible and Tyson2007112Tyson et al.2019, this formalism does not scale up well to large signalling networks, due to the difficulty of estimating kinetic parameter values 6Babtie and Stumpf2017. In contrast, the logical (or logic) modelling formalism represents a simpler means of abstraction where the causal relationships between proteins (or genes) are encoded with logic statements, and dynamical behaviours are represented by transitions between discrete states of the system 57Kauffman1969107Thomas1973. In particular, Boolean models, the simplest implementation of logical models, describe each protein as a binary variable (ON/OFF). This framework is flexible, requires in principle no quantitative information, can be hence applied to large networks combining multiple pathways, and can also provide a qualitative understanding of molecular systems lacking detailed mechanistic information.
In the last years, logical and, in particular, Boolean modelling has been successfully used to describe the dynamics of human cellular signal transduction and gene regulations 13Calzone et al.201021Cho et al.201635Flobak et al.201546Grieco et al.201348Helikar et al.2008109Traynard et al.2016 and their deregulation in cancer 39Fumiã and Martins201352Hu et al.2015. Numerous applications of logical modelling have shown that this framework is able to delineate the main dynamical properties of complex biological regulatory networks 1Abou-Jaoudé et al.201134Fauré et al.2006.
However, the Boolean approach is purely qualitative and does not consider the real time of cellular events (half time of proteins, triggering of apoptosis, etc.). To cope with this issue, we developed the MaBoSS software to compute continuous Markov Chain simulations on the model state transition graph (STG), in which a model state is defined as a vector of nodes that are either active or inactive. In practice, MaBoSS associates transition rates for activation and inhibition of each node of the network, enabling it to account for different time scales of the processes described by the model. Given some initial conditions, MaBoSS applies a Monte-Carlo kinetic algorithm (or Gillespie algorithm) to the STG to produce time trajectories 104Stoll et al.2017103Stoll et al.2012 such that the time evolution of the model state probabilities can be estimated. Stochastic simulations can easily explore the model dynamics with different initial conditions by varying the probability of having a node active at the beginning of the simulations and by modifying the model such that it accounts for genetic and environmental perturbations (e.g. presence or absence of growth factors or death receptors). For each case, the effect on the probabilities of selected read-outs can be measured 23Cohen et al.201577Montagud et al.2019.
When summarising the biological knowledge into a network and translating it into logical terms, the obtained model is generic and cannot explain the differences and heterogeneity between patients’ responses to treatments. Models can be trained with dedicated perturbation experiments 30Dorier et al.201693Saez-Rodriguez et al.2009, but such data can only be obtained with non-standard procedures such as microfluidics from patients’ material 32Eduati et al.2020. To address this limitation, we developed a methodology to use different omics data that are more commonly available to personalise generic models to individual cancer patients or cell lines and verified that the obtained models correlated with clinical results such as patient survival information 9Béal et al.2019. In the present work, we apply this approach to prostate cancer to suggest targeted therapy to patients based on their omics profile (Figure 1). We first built 488 patient- and eight cell line prostate-specific models using data from The Cancer Genome Atlas (TCGA) and the Genomics of Drug Sensitivity in Cancer (GDSC) projects, respectively. Simulating these models with the MaBoSS framework, we identified points of intervention that diminish the probability of reaching pro-tumorigenic phenotypes. Lastly, we developed a new methodology to simulate drug effects on these data-tailored Boolean models and present a list of viable drugs and treatments that could be used on these patient- and cell line-specific models for optimal results. Experimental validations were performed on the LNCaP prostate cell line with two predicted targets, confirming the predictions of the model.

Workflow to build patient-specific Boolean models and to uncover personalised drug treatments from present work.
We gathered data from 39Fumiã and Martins2013 Boolean model, Omnipath 111Türei et al.2021 and pathways identified with ROMA 74Martignetti et al.2016 on the TCGA data to build a prostate-specific prior knowledge network. This network was manually converted into a prostate Boolean model that could be stochastically simulated using MaBoSS 104Stoll et al.2017 and tailored to different TCGA and GDSC datasets using our PROFILE tool to have personalised Boolean models. Then, we studied all the possible single and double mutants on these tailored models using our logical pipeline of tools 77Montagud et al.2019. Using these personalised models and our PROFILE_v2 tool presented in this work, we obtained tailored drug simulations and drug treatments for 488 TCGA patients and eight prostate cell lines. Lastly, we performed drug-dose experiments on a shortlist of candidate drugs that were particularly interesting in the LNCaP prostate cell line. Created with BioRender.com.
Results
Prostate Boolean model construction
A network of signalling pathways and genes relevant for prostate cancer progression was assembled to recapitulate the potential deregulations that lead to high-grade tumours. Dynamical properties were added onto this network to perform simulations, uncover therapeutic targets and explore drug combinations. The model was built upon a generic cancer Boolean model by 39Fumiã and Martins2013, which integrates major signalling pathways and their substantial cross-talks. The pathways include the regulation of cell death and proliferation in many tumours.
This initial generic network was extended to include prostate cancer-specific genes (e.g. SPOP, AR, etc.), pathways identified using ROMA 74Martignetti et al.2016, OmniPath 111Türei et al.2021, and up-to-date literature. ROMA is applied on omics data, either transcriptomics or proteomics. In each pathway, the genes that contribute the most to the overdispersion are selected. ROMA was applied to the TCGA transcriptomics data using gene sets from cancer pathway databases (Appendix 1, Section 1.1.3, Appendix 1—figure 1). These results were used as guidelines to extend the network to fully cover the alterations found in prostate cancer patients. OmniPath was used to complete our network finding connections between the proteins of interest known to play a role in the prostate and the ones identified with ROMA, and the list of genes already present in the model (Appendix 1, Sections 1.1.3 and 1.1.4, Appendix 1—figures 2 and 3). The final network includes pathways such as androgen receptor, MAPK, Wnt, NFkB, PI3K/AKT, MAPK, mTOR, SHH, the cell cycle, the epithelial-mesenchymal transition (EMT), apoptosis and DNA damage pathways.
This network was then converted into a Boolean model where variables can take two values: 0 (inactivate or absent) or 1 (activate or present). Our model aims at predicting prostate phenotypic behaviours for healthy and cancer cells in different conditions. Nine inputs that represent some of these physiological conditions of interest were considered: Epithelial Growth Factor (EGF), Fibroblast Growth Factor (FGF), Transforming Growth Factor beta (TGFbeta), Nutrients, Hypoxia, Acidosis, Androgen, Tumour Necrosis Factor alpha (TNF alpha), and Carcinogen. These input nodes have no regulation. Their value is fixed according to the simulated experiment to represent the status of the microenvironmental characteristics (e.g. the presence or absence of growth factors, oxygen, etc.). A more complex multiscale approach would be required to consider the dynamical interaction with other cell types and the environment.
We defined six variables as output nodes that allow the integration of multiple phenotypic signals and simplify the analysis of the model. Two of these phenotypes represent the possible growth status of the cell: Proliferation and Apoptosis. Apoptosis is activated by Caspase 8 or Caspase 9, while Proliferation is activated by cyclins D and B (read-outs of the G1 and M phases, respectively). The Proliferation output is described in published models as specific stationary protein activation patterns, namely the following sequence of activation of cyclins: Cyclin D, then Cyclin E, then Cyclin A, and finally Cyclin B 109Traynard et al.2016. Here, we considered a proper sequence when Cyclin D activates first, allowing the release of the transcriptional factor E2F1 from the inhibitory complex it was forming with the RB (retinoblastoma protein), and then triggering a series of events leading to the activation of Cyclin B, responsible for the cell’s entry into mitosis (Appendix 1, Section 2.2, Appendix 1—figure 5). We also define several phenotypic outputs that are readouts of cancer hallmarks: Invasion, Migration, (bone) Metastasis and DNA repair. The final model accounts for 133 nodes and 449 edges (Figure 2, Supplementary file 1, and in GINsim format at the address: http://ginsim.org/model/signalling-prostate-cancer).
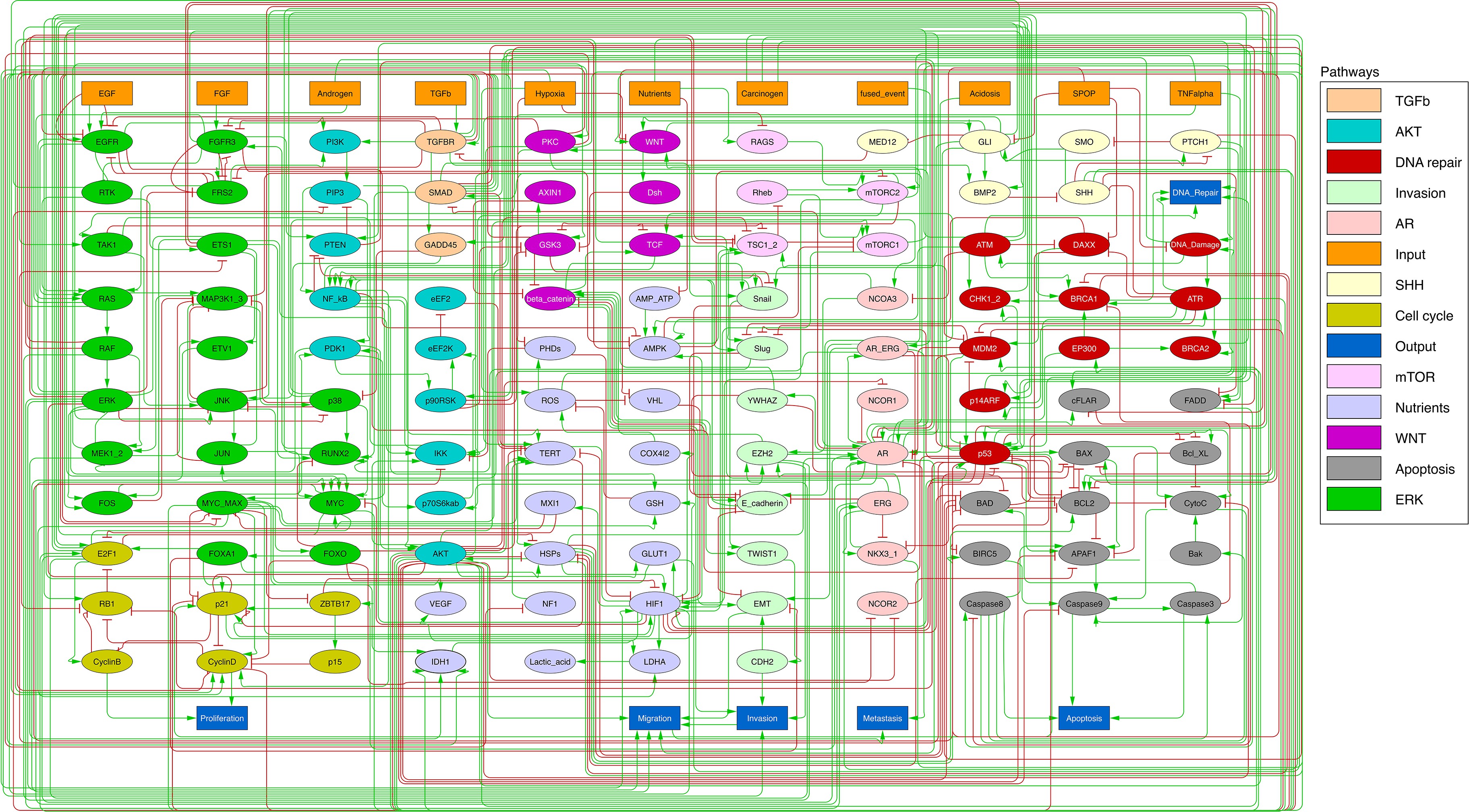
Prostate Boolean model used in present work.
Nodes (ellipses) represent biological entities, and arcs are positive (green) or negative (red) influences of one entity on another one. Orange rectangles correspond to inputs (from left to right: Epithelial Growth Factor (EGF), Fibroblast Growth Factor (FGF), Transforming Growth Factor beta (TGFbeta), Nutrients, Hypoxia, Acidosis, Androgen, fused_event, Tumour Necrosis Factor alpha (TNFalpha), SPOP, Carcinogen) and dark blue rectangles to outputs that represent biological phenotypes (from left to right: Proliferation, Migration, Invasion, Metastasis, Apoptosis, DNA_repair), the read-outs of the model. This network is available to be inspected as a Cytoscape file in the Supplementary file 1.
Prostate Boolean model simulation
The model can be considered as a model of healthy prostate cells when no mutants (or fused genes) are present. We refer to this model as the wild type model. These healthy cells mostly exhibit quiescence (neither proliferation nor apoptosis) in the absence of any input (Figure 3A). When Nutrients and growth factors (EGF or FGF) are present, Proliferation is activated (Figure 3B). Androgen is necessary for AR activation and helps in the activation of Proliferation, even though it is not necessary when Nutrients or growth factors are present. Cell death factors (such as Caspase 8 or 9) trigger Apoptosis in the absence of SPOP, while Hypoxia and Carcinogen facilitate apoptosis but are not necessary if cell death factors are present (Figure 3C).

Prostate Boolean model MaBoSS simulations.
(A) The model was simulated with all initial inputs set to 0 and all other variables random. All phenotypes are 0 at the end of the simulations, which should be understood as a quiescent state, where neither proliferation nor apoptosis is active. (B) The model was simulated with growth factors (EGF and FGF), Nutrients and Androgen ON. (C) The model was simulated with Carcinogen, Androgen, TNFalpha, Acidosis, and Hypoxia ON.
In our model, the progression towards metastasis is described as a stepwise process. Invasion is first activated by known pro-invasive proteins: either β-catenin 38Francis et al.2013 or a combination of CDH2 29De Wever et al.2004, SMAD 27Daroqui et al.2012, or EZH2 88Ren et al.2012. Migration is then activated by Invasion and EMT and with either AKT or AR 17Castoria et al.2011. Lastly, (bone) Metastasis is activated by Migration and one of three nodes: RUNX2 5Altieri et al.2009, ERG 3Adamo and Ladomery2016 or ERG fused with TMPRSS2 102St John et al.2012, FLI1, ETV1 or ETV4 15Cancer Genome Atlas Research Network2015.
This prostate Boolean model was simulated stochastically using MaBoSS 104Stoll et al.2017103Stoll et al.2012 and validated by recapitulating known phenotypes of prostate cells under physiological conditions (Figure 3 and Appendix 1, Sections 2.2 and 2.3, Appendix 1—figures 5–7). In particular, we tested that combinations of inputs lead to non-aberrant phenotypes such as growth factors leading to apoptosis in wild type conditions; we also verified that the cell cycle events occur in proper order: as CyclinD gets activated, RB1 is phosphorylated and turned OFF, allowing E2F1 to mediate the synthesis of CyclinB (see Supplementary file 2 for the jupyter notebook and the simulation of diverse cellular conditions).
Personalisation of the prostate Boolean model
Personalised TCGA prostate cancer patient Boolean models
We tailored the generic prostate Boolean model to a set of 488 TCGA prostate cancer patients (Appendix 1, Section 4, Appendix 1—figure 9) using our personalisation method (PROFILE) 9Béal et al.2019, constructing 488 individual Boolean models, one for each patient. Personalised models were built using three types of data: discrete data such as mutations and copy number alterations (CNA) and continuous data such as RNAseq data. For discrete data, the nodes corresponding to the mutations or the CNA were forced to 0 or 1 according to the effect of alterations, based on a priori knowledge (i.e. if the mutation was reported to be activating or inhibiting the gene’s activity). For continuous data, the personalisation method modifies the value for the transition rates of model variables and their initial conditions to influence the probability of some transitions. This corresponds, in a biologically meaningful way, to translating genetic mutations as lasting modifications making the gene independent of regulation, and to translating RNA expression levels as modulation of a signal but not changing the regulation rules (see Materials and methods and in Appendix 1, Section 4.1, Appendix 1—figures 10–14).
We assess the general behaviour of the individual patient-specific models by comparing the model outputs (i.e. probabilities to reach certain phenotypes) with clinical data. Here, the clinical data consist of a Gleason grade score associated with each patient, which in turn corresponds to the gravity of the tumour based on its appearance and the stage of invasion 19Chen and Zhou201643Gleason199242Gleason and Tannenbaum1977. We gathered the output probabilities for all patient-specific models and confronted them to their Gleason scores. The phenotype DNA_repair, which can be interpreted as a sensor of DNA damage and genome integrity which could lead to DNA repair, seems to separate low and high Gleason scores (Figure 4A and Appendix 1, Section 4.1, Appendix 1—figures 15–18), confirming that DNA damage pathways are activated in patients 73Marshall et al.2019 but may not lead to the triggering of apoptosis in this model (Appendix 1, Section 4.1, Appendix 1—figure 11). Also, the centroids of Gleason grades tend to move following Proliferation, Migration and Invasion variables. We then looked at the profiles of the phenotype scores across patients and their Gleason grade and found that the density of high Proliferation score (close to 1, Figure 4B) tends to increase as the Gleason score increases (from low to intermediate to high) and these distributions are significantly different (Kruskal-Wallis rank sum test, p-value = 0.00207; Appendix 1, Section 4.1). The Apoptosis phenotype probabilities, however, do not have a clear trend across grades (Figure 4C), even though the distributions are significantly different (Kruskal-Wallis rank sum test, p-value = 2.83E-6; Appendix 1, Section 4.1).
# Load packages
list.of.packages <- c("ggplot2", "tidyverse", "gghalves","patchwork", "factoextra", "ggpubr","ggsci")
pacman::p_load(list.of.packages, character.only = TRUE)
load(url("https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20TCGA%20patients'%20simulations/data_plot_TCGA.RData.txt"))
load(url("https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20TCGA%20patients'%20simulations/res.pca.RData.txt"))
coloursG3 <- c("Low" = "yellowgreen", "Interm" = "orange3", "High" = "black")
r <- 7.16
Associations between simulations and Gleason grades (GG).
(A) Centroids of the Principal Component Analysis of the samples according to their Gleason grades (GG). The personalisation recipe used was mutations and copy number alterations (CNA) as discrete data and RNAseq as continuous data. Density plots of Proliferation (B) and Apoptosis (C) scores according to GG; each vignette corresponds to a specific sub-cohort with a given GG. Kruskal-Wallis rank sum test across GG is significant for Proliferation (p-value = 0.00207) and Apoptosis (p-value = 2.83E-6).
R code needed to obtain Figure 4.Processed datasets needed are Figure 4—source data 1 and Figure 4—source data 2 are located in the corresponding folder of the repository: here.Processed dataset needed to obtain the phenotype distributions of Figure 4B, C, with Figure 4—source code 1.Processed dataset needed to obtain the PCA of Figure 4A, with Figure 4—source code 1.
chunk: Figure 4
p_prolif <- ggplot(data_plot_TCGA, aes(x=Proliferation, fill=GG3)) +
geom_density(show.legend = FALSE) +
facet_grid(GG3~.) +
theme_bw()+
scale_fill_manual(values=coloursG3)
p_apoptosis <- ggplot(data_plot_TCGA, aes(x=Apoptosis, fill=GG3)) +
geom_density() +
facet_grid(GG3~.) +
scale_fill_manual(values=coloursG3,
guide = guide_legend(direction = "vertical")) +
theme_bw() +
labs(fill="Gleason\ngroups")
data_plot_3 <- filter(data_plot_TCGA, !is.na(GG3)) %>%
group_by(GG3) %>%
summarise(Dim.1=mean(Dim.1, na.rm=T),Dim.2=mean(Dim.2, na.rm=T))
p_bary3 <- fviz_pca_var(res.pca,scale. = r/5, repel = T,
select.var = list(name=c("Proliferation", "Apoptosis",
"DNA_Repair", "Migration",
"Invasion"))) +
geom_point(data = data_plot_3, aes(x=Dim.1, y=Dim.2, color=GG3),
size=3, show.legend = FALSE
) +
scale_color_manual(values=coloursG3, name="GG\nBarycent.") +
theme_bw() +
labs(title="")
t <- (p_bary3 / (p_prolif | p_apoptosis) | guide_area()) +
plot_layout(guides = 'collect', widths = c(6,1)) +
plot_annotation(tag_level=c('A', 'B', 'C')) +
theme_pubclean() +
theme(plot.tag = element_text(size = 18))
t
::: {figalign="center"}
::: {#fig4}
Personalised drug predictions of TCGA Boolean models
Using the 488 TCGA patient-specific models, we looked in each patient for genes that, when inhibited, hamper Proliferation or promote Apoptosis in the model. We focused on these inhibitions as most drugs interfere with the protein activity related to these genes, even though our methodology allows us to study increased protein activity related to over-expression of genes as well 9Béal et al.201977Montagud et al.2019. Interestingly, we found several genes that were found as suitable points of intervention in most of the patients (MYC_MAX complex and SPOP were identified in more than 80% of the cases) (Appendix 1, Section 4.2, Appendix 1—figures 19 and 20), but others were specific to only some of the patients (MXI1 was identified in only 4 patients, 1% of the total, GLI in only 7% and WNT in 8% of patients). All the TCGA-specific personalised models can be found in Supplementary file 3, and the TCGA mutants and their phenotype scores can be found in Supplementary file 4.
Furthermore, we explored the possibility of finding combinations of treatments that could reduce the Proliferation phenotype or increase the Apoptosis one. To lower the computational power need, we narrowed down the list of potential candidates to a set of selected genes that are targets of already-developed drugs relevant in cancer progression (Table 1) and analysed the simulations of the models with all the single and combined perturbations.
| Node | Gene | Compound / Inhibitor name | Clinical stage | Source |
|---|---|---|---|---|
| AKT | AKT1, AKT2, AKT3 | PI-103 | Preclinical | Drug Bank |
| Enzastaurin | Phase 3 | Drug Bank | ||
| Archexin, Pictilisib | Phase 2 | Drug Bank | ||
| AR | AR | Abiraterone,Enzalutamide, Formestane, Testosterone propionate | Approved | Drug Bank |
| 5alpha-androstan-3beta-ol | Preclinical | Drug Bank | ||
| Caspase8 | CASP8 | Bardoxolone | Preclinical | Drug Bank |
| cFLAR | CFLAR | - | - | - |
| EGFR | EGFR | Afatinib, Osimertinib, Neratinib, Erlotinib, Gefitinib | Approved | Drug Bank |
| Varlitinib | Phase 3 | Drug Bank | ||
| Olmutinib, Pelitinib | Phase 2 | Drug Bank | ||
| ERK | MAPK1 | Isoprenaline | Approved | Drug Bank |
| Perifosine | Phase 3 | Drug Bank | ||
| Turpentine, SB220025, Olomoucine, Phosphonothreonine | Preclinical | Drug Bank | ||
| MAPK3, MAPK1 | Arsenic trioxide | Approved | Drug Bank | |
| Ulixertinib, Seliciclib | Phase 2 | Drug Bank | ||
| Purvalanol | Preclinical | Drug Bank | ||
| MAPK3 | Sulindac, Cholecystokinin | Approved | Drug Bank | |
| 5-iodotubercidin | Preclinical | Drug Bank | ||
| GLUT1 | SLC2A1 | Resveratrol | Phase 4 | Drug Bank |
| HIF-1 | HIF1A | CAY-10585 | Preclinical | Drug Bank |
| HSPs | HSP90AA1, HSP90AB1, HSP90B1, HSPA1A, HSPA1B, HSPB1 | Cladribine | Approved | Drug Bank |
| 17-DMAG | Phase 2 | Drug Bank | ||
| NMS-E973 | Preclinical | Drug Bank | ||
| MEK1_2 | MAP2K1, MAP2K2 | Trametinib, Selumetinib | Approved | Drug Bank |
| Perifosine | Phase 3 | Drug Bank | ||
| PD184352 (CI-1040) | Phase 2 | Drug Bank | ||
| MYC_MAX | complex of MYC and MAX | 10058-F4 (for MAX) | Preclinical | Drug Bank |
| p14ARF | CDKN2A | - | - | - |
| PI3K | PIK3CA, PIK3CB, PIK3CG, PIK3CD, PIK3R1, PIK3R2, PIK3R3, PIK3R4, PIK3R5, PIK3R6, PIK3C2A, PIK3C2B, PIK3C2G, PIK3C3 | PI-103 | Preclinical | Drug Bank |
| Pictilisib | Phase 2 | Drug Bank | ||
| ROS | NOX1, NOX3, NOX4 | Fostamatinib | Approved | Drug Bank |
| NOX2 | Dextromethorphan | Approved | Drug Bank | |
| Tetrahydroisoquinolines (CHEMBL3733336, CHEMBL3347550, CHEMBL3347551) | Preclinical | ChEMBL | ||
| SPOP | SPOP | - | - | - |
| TERT | TERT | Grn163l | Phase 2 | Drug Bank |
| BIBR 1532 | Preclinical | ChEMBL |
We used the models to grade the effect that the combined treatments have in each one of the 488 TCGA patient-specific models’ phenotypes. This list of combinations of treatments can be used to compare the effects of drugs on each TCGA patient and allows us to propose some of them for individual patients and to suggest drugs suitable to groups of patients (Supplementary file 4). Indeed, the inactivation of some of the targeted genes had a greater effect in some patients than in others, suggesting the possibility for the design of personalised drug treatments. For instance, for the TCGA-EJ-5527 patient, the use of MYCMAX complex inhibitor reduced _Proliferation to 66%. For this patient, combining MYCMAX with other inhibitors, such as AR or AKT, did not further reduce the _Proliferation score (67% in these cases). Other patients have MYCMAX as an interesting drug target, but the inhibition of this complex did not have such a dramatic effect on their _Proliferation scores as in the case of TCGA-EJ-5527. Likewise, for the TCGA-H9-A6BX patient, the use of SPOP inhibitor increased Apoptosis by 87%, while the use of a combination of cFLAR and SPOP inhibitors further increased Apoptosis by 89%. For the rest of this section, we focus on the analysis of clinical groups rather than individuals.
Studying the decrease of Proliferation, we found that AKT is the top hit in Gleason Grades 1, 2, 3, and 4, seconded by EGFR and SPOP in Grade 1, by SPOP and PIP3 in Grade 2, by PIP3 and AR in Grade 3, and by CyclinD and MYCMAX in Grade 4. MYC_MAX is the top hit in Grade 5, seconded by AR (Appendix 1, Section 4.2, Appendix 1—figure 19). In regard to the increase of _Apoptosis, SPOP is the top hit in all grades, seconded by SSH in Grades 1, 2, and 3 and by AKT in Grade 4 (Appendix 1, Section 4.2, Appendix 1—figure 20). It is interesting to note here that many of these genes are targeted by drugs (Table 1). Notably, AR is the target of the drug Enzalutamide, which is indicated for men with an advanced stage of the disease 97Scott2018, or that MYC is the target of BET bromodomain inhibitors and are generally effective in castration-resistant prostate cancer cases 24Coleman et al.2019.
The work on patient data provided possible insights and suggested patient- and grade-specific potential targets. To validate our approach experimentally, we personalised the prostate model to different prostate cell lines, where we performed drug assays to confirm the predictions of the model.
Personalised drug predictions of LNCaP Boolean model
We applied the methodology for personalisation of the prostate model to eight prostate cell lines available in GDSC 53Iorio et al.2016: 22RV1, BPH-1, DU-145, NCI-H660, PC-3, PWR-1E, and VCaP (results in Appendix 1, Section 5 and are publicly available in Supplementary file 5). We decided to focus the validation on one cell line, LNCaP.
LNCaP, first isolated from a human metastatic prostate adenocarcinoma found in a lymph node 51Horoszewicz et al.1983, is one of the most widely used cell lines for prostate cancer studies. Androgen-sensitive LNCaP cells are representative of patients sensitive to treatments as opposed to resistant cell lines such as DU-145. Additionally, LNCaP cells have been used to obtain numerous subsequent derivatives with different characteristics 26Cunningham and You2015.
The LNCaP personalisation was performed based on mutations as discrete data and RNA-Seq as continuous data. The resulting LNCaP-specific Boolean model was then used to identify all possible combinations of mutations (interpreted as effects of therapies) and to study the synergy of these perturbations. For that purpose, we automatically performed single and double mutant analyses on the LNCaP-specific model (knock-out and overexpression) 77Montagud et al.2019 and focused on the model phenotype probabilities as read-outs of the simulations. The analysis of the complete set of simulations for the 32,258 mutants can be found in the Appendix 1, Section 6.1 and in Supplementary file 6, where the LNCaP cell line-specific mutants and their phenotype scores are reported for all mutants. Among all combinations, we identified the top 20 knock-out mutations that depleted Proliferation or increased Apoptosis the most. As some of them overlapped, we ended up with 29 nodes: AKT, AR, ATR, AXIN1, Bak, BIRC5, CDH2, cFLAR, CyclinB, CyclinD, E2F1, eEF2K, eEF2, eEF2K, EGFR, ERK, HSPs, MED12, mTORC1, mTORC2, MYC, MYC_MAX, PHDs, PI3K, PIP3, SPOP, TAK1, TWIST1, and VHL. We used the scores of these nodes to further trim down the list to have 10 final nodes (AKT, AR, cFLAR, EGFR, ERK, HSPs, MYC_MAX, SPOP, and PI3K) and added seven other nodes whose genes are considered relevant in cancer biology, such as AR_ERG fusion, Caspase8, HIF1, GLUT1, MEK1_2, p14ARF, ROS, and TERT (Table 1). We did not consider the overexpression mutants as they have a very difficult translation to drug uses and clinical practices.
To further analyse the mutant effects, we simulated the LNCaP model with increasing node inhibition values to mimic the effect of drugs’ dosages using a methodology we specifically developed for this purpose (PROFILEv2 and available at https://github.com/ArnauMontagud/PROFILE_v2; 79Montagud2022). Six simulations were done for each inhibited node, with 100% of node inhibition (proper knock-out), 80%, 60%, 40%, 20% and 0% (no inhibition) (see Materials and methods). A nutrient-rich media with EGF was used for these simulations that correspond to experimental conditions that are tested here. We show results on three additional sets of initial conditions in the Appendix 1, Section 6, Appendix 1—figure 27: a nutrient-rich media with androgen, with androgen and EGF, and with none, . We applied this gradual inhibition, using increasing drugs’ concentrations, to a reduced list of drug-targeted genes relevant for cancer progression (Table 1). We confirmed that the inhibition of different nodes affected differently the probabilities of the outputs (Appendix 1, Section 7.3.1, Appendix 1—figures 34 and 35). Notably, the _Apoptosis score was slightly promoted when knocking out SPOP under all growth conditions (Appendix 1, Section 7.3.1, Appendix 1—figure 35). Likewise, Proliferation depletion was accomplished when HSPs or MYC_MAX were inhibited under all conditions and, less notably, when ERK, EGFR, SPOP, or PI3K were inhibited (Appendix 1, Section 7.3.1, Appendix 1—figure 35).
Additionally, these gradual inhibition analyses can be combined to study the interaction of two simultaneously inhibiting nodes (Appendix 1, Section 7.3.2, Appendix 1—figures 36 and 37). For instance, the combined gradual inhibition of ERK and MYC_MAX nodes affects the Proliferation score in a balanced manner (Figure 5A) even though MYC_MAX seems to affect this phenotype more, notably at low activity levels. By extracting subnetworks of interaction around ERK and MYC_MAX and comparing them, we found that the pathways they belong to have complementary downstream targets participating in cell proliferation through targets in MAPK and cell cycle pathways. This complementarity could explain the synergistic effects observed (Figure 5A and C).
Phenotype score variations and synergy upon combined ERK and MYCMAX (A and C) and HSPs and PI3K (B and D) inhibition under _EGF growth condition.
Proliferation score variation (A) and Bliss Independence synergy score (C) with increased node activation of nodes ERK and MYC_MAX. Proliferation score variation (B) and Bliss Independence synergy score (D) with increased node activation of nodes HSPs and PI3K. Bliss Independence synergy score <1 is characteristic of drug synergy, grey colour means one of the drugs is absent and thus no synergy score is available.
R code needed to perform the drug dosage experiments and obtain Figure 5 from the main text and Appendix 1—figures 27, 34–39.Processed datasets needed is Figure 5—source data 1 and is located in the corresponding folder of the repository: here.Processed datasets needed to obtain the phenotype score variations and synergy values of Figure 5 with Figure 5—source code 1.
chunk: Figure 5
load(url("https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Gradient%20inhibition%20of%20nodes/drugs_figures_datasets.RData.txt"))
interesants_prolif <-
tot %>% filter(drug1 == "ERK" & drug2 == "MYC_MAX")
interesants_bliss <-
tot_Bliss_prolif %>% filter(drug1 == "ERK" & drug2 == "MYC_MAX")
prolif <-
ggplot(interesants_prolif %>% filter(Phenotype == "Proliferation"),
aes(dose1, dose2)) +
geom_tile(aes(fill = value), colour = "black") +
facet_grid(drug2 ~ drug1, switch = "both") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradient2(low = "blue",
high = "white",
limits = c(-1, 0.1)) +
labs(x = "ERK node inhibition (%)",
y = "MYC_MAX node\ninhibition (%)",
fill = "Treated -\nuntreated\ncell line\nscore") +
theme(
legend.position = "none",
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.text = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5)
)
prolifBliss <- ggplot(interesants_bliss, aes(dose1, dose2)) +
geom_tile(aes(fill = Bliss), colour = "black") +
facet_grid(drug2 ~ drug1, switch = "both") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradientn(
colours = c("#8c5ba9", "white", "#7fbf7b"),
breaks = (c(0, 0.5, 1, 1.5, 2)),
labels = (c(0, 0.5, 1, 1.5, ">2")),
limits = c(0, 2)
) +
labs(x = "ERK node inhibition (%)",
y = "MYC_MAX node\ninhibition (%)",
fill = "Bliss\nscore") +
theme(
legend.position = "none",
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.text = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5)
)
interesants_prolif <-
tot %>% filter(drug1 == "HSPs" & drug2 == "PI3K")
interesants_bliss <-
tot_Bliss_prolif %>% filter(drug1 == "HSPs" & drug2 == "PI3K")
prolif2 <-
ggplot(interesants_prolif %>% filter(Phenotype == "Proliferation"),
aes(dose1, dose2)) +
geom_tile(aes(fill = value), colour = "black") +
facet_grid(drug2 ~ drug1, switch = "both") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradient2(low = "blue",
high = "white",
limits = c(-1, 0.1)) +
labs(x = "HSPs node inhibition (%)",
y = "PI3K node inhibition (%)",
fill = "Treated -\nuntreated\ncell line\nscore") +
theme(
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.text = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5)
)
prolifBliss1 <- ggplot(interesants_bliss, aes(dose1, dose2)) +
geom_tile(aes(fill = Bliss), colour = "black") +
facet_grid(drug2 ~ drug1, switch = "both") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradientn(
colours = c("#8c5ba9", "white", "#7fbf7b"),
breaks = (c(0, 0.5, 1, 1.5, 2)),
labels = (c(0, 0.5, 1, 1.5, ">2")),
limits = c(0, 2)
) +
labs(x = "HSPs node inhibition (%)",
y = "PI3K node inhibition (%)",
fill = "Bliss\nscore") +
theme(
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.text = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5)
)
prolif + prolif2 + prolifBliss + prolifBliss1 + plot_annotation(tag_levels = 'A')
::: {figalign="center"}
::: {#fig5}
Proliferation phenotype score variations of the LNCaP model upon combined nodes inhibition under EGF growth condition.
Figure 5A is a closer look at ERK and MYC_MAX combination and Figure 5B at HSPs and PI3K combination.
chunk: Figure S36
# Figure S36: Double drug Proliferation scores
Double_EGF_Proliferation <- ggplot(
tot %>% filter(Phenotype == "Proliferation"), aes(dose1, dose2)) +
facet_grid(drug2 ~ drug1, switch = "both") +
geom_tile(aes(fill = value), colour = "black") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradient2(
low = "blue",
high = "firebrick",
mid = "white",
limits = c(-1, 1)
) +
labs(
title = paste0("Proliferation phenotype variations upon drugs inhibition"),
x = "Node inhibition (%)",
y = "Node inhibition (%)",
fill = "Treated -\nuntreated\ncell line\nscore"
) +
theme(
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5),
strip.text.y.left = element_text(angle = 0),
strip.text.x = element_text(angle = 90),
axis.text.x = element_text(angle = 90)
)
Double_EGF_Proliferation
::: {figalign="center"}
::: {#app1fig36}
Apoptosis phenotype score variations of the LNCaP model upon combined nodes inhibition under EGF growth condition.
chunk: Figure S37
# Figure S37: Double drug Apoptosis scores
Double_EGF_Apoptosis <- ggplot(
tot %>% filter(Phenotype == "Apoptosis"), aes(dose1, dose2)) +
facet_grid(drug2 ~ drug1, switch = "both") +
geom_tile(aes(fill = value), colour = "black") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradient2(low = "blue", high = "firebrick", mid = "white") +
labs(
title = paste0("Apoptosis phenotype variations upon drugs inhibition"),
x = "Node inhibition (%)",
y = "Node inhibition (%)",
fill = "Treated -\nuntreated\ncell line\nscore"
) +
theme(
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5),
strip.text.y.left = element_text(angle = 0),
strip.text.x = element_text(angle = 90),
axis.text.x = element_text(angle = 90)
)
Double_EGF_Apoptosis
::: {figalign="center"}
::: {#app1fig37}
Bliss Independence synergies scores variations in Proliferation phenotype of the LNCaP model upon combined nodes inhibition under EGF growth conditions.
Bliss Independence synergy score <1 is characteristic of drug synergy. Figure 5C is a closer look at ERK and MYC_MAX combination and Figure 5D at HSPs and PI3K combination, grey colour means one of the drugs is absent and thus no synergy score is available.
chunk: Figure S38
# Figure S38: Double drug Proliferation Bliss synergy values
bliss_pro <- ggplot(tot_Bliss_prolif, aes(dose1, dose2)) +
geom_tile(aes(fill = Bliss), colour = "black") +
facet_grid(drug2 ~ drug1, switch = "both") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradientn(
colours = c("#8c5ba9", "white", "#7fbf7b"),
breaks = (c(0.0, 0.5, 1, 1.5, 2)),
labels = (c(0.0, 0.5, 1, 1.5, ">2"))
) +
labs(x = "Node inhibition (%)",
y = "Node inhibition (%)",
fill = "Bliss score") +
theme(
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5),
strip.text.y.left = element_text(angle = 0),
strip.text.x = element_text(angle = 90),
axis.text.x = element_text(angle = 90)
)
bliss_pro
::: {figalign="center"}
::: {#app1fig38}
Bliss Independence synergies scores variations in Apoptosis phenotypes of the LNCaP model upon combined nodes inhibition under EGF growth conditions.
Bliss Independence synergy score <1 is characteristic of drug synergy, grey colour means one of the drugs is absent and thus no synergy score is available.
chunk: Figure S39
# Figure S39: Double drug Apoptosis Bliss synergy values
bliss_apop <- ggplot(tot_Bliss_apop, aes(dose1, dose2)) +
geom_tile(aes(fill = Bliss), colour = "black") +
facet_grid(drug2 ~ drug1, switch = "both") +
scale_x_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_y_continuous(
breaks = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
labels = c(0, 20, 40, 60, 80, 100)
) +
scale_fill_gradientn(
colours = c("#8c5ba9", "white", "#7fbf7b"),
breaks = (c(0, 0.5, 1, 1.5, 2)),
labels = (c(0, 0.5, 1, 1.5, ">2"))
) +
labs(x = "Node inhibition (%)",
y = "Node inhibition (%)",
fill = "Bliss score") +
theme(
axis.ticks = element_blank(),
panel.background = element_blank(),
strip.background = element_rect(fill = NA),
plot.title = element_text(hjust = 0.5),
strip.text.y.left = element_text(angle = 0),
strip.text.x = element_text(angle = 90),
axis.text.x = element_text(angle = 90)
)
bliss_apop
::: {figalign="center"}
::: {#app1fig39}
Lastly, drug synergies can be studied using Bliss Independence using the results from single and combined simulations with gradual inhibitions. This score compares the combined effect of two drugs with the effect of each one of them, with a synergy when the value of this score is lower than 1. We found that the combined inhibition of ERK and MYC_MAX nodes on the Proliferation score was synergistic (Figure 5C). Another synergistic pair is the combined gradual inhibition of HSPs and PI3K nodes that also affects the Proliferation score in a joint manner (Figure 5B), with some Bliss Independence synergy found (Figure 5D). A complete study on the Bliss Independence synergy of all the drugs considered in the present work on Proliferation and Apoptosis phenotypes can be found in Appendix 1, Section 7.3.2, Appendix 1—figures 38 and 39.
Experimental validation of predicted targets
Drugs associated with the proposed targets
To identify drugs that could act as potential inhibitors of the genes identified with the Boolean model, we explored the drug-target associations in DrugBank 115Wishart et al.2018 and ChEMBL 40Gaulton et al.2017. We found drugs that targeted almost all genes corresponding to the nodes of interest in Table 1, except for cFLAR, p14ARF, and SPOP. However, we could not identify experimental cases where drugs targeting both members of the proposed combinations were available (Appendix 1, Section 7.1 and in Supplementary file 6). One possible explanation is that the combinations predicted by the model suggest, in some cases, to overexpress the potential target and most of the drugs available act as inhibitors of their targets.
Using the cell line-specific models, we tested if the LNCaP cell line was more sensitive than the rest of the prostate cell lines to the LNCaP-specific drugs identified in Table 1. We compared GDSC’s Z-score of these drugs in LNCaP with their Z-scores in all GDSC cell lines (Figure 6 and Appendix 1, Section 7.2, Appendix 1—figure 33). We observed that LNCaP is more sensitive to drugs targeting AKT or TERT than the rest of the studied prostate cell lines. Furthermore, we saw that the drugs that targeted the genes included in the model allowed the identification of cell line specificities (Appendix 1, Section 7.1). For instance, target enrichment analysis showed that LNCaP cell lines are especially sensitive to drugs targeting PI3K/AKT/mTOR, hormone-related (AR targeting) and Chromatin (bromodomain inhibitors, regulating Myc) pathways (adjusted p-values from target enrichment: 0.001, 0.001, and 0.032, respectively, Appendix 1, Section 7.1, Appendix 1—table 2), which corresponds to the model predictions (Table 1). Also, the LNCaP cell line is more sensitive to drugs targeting model-identified nodes than to drugs targeting other proteins (Appendix 1, Section 7.1, Appendix 1—figure 32, Mann-Whitney U p-value 0.00041), and this effect is specific for LNCaP cell line (Mann-Whitney U p-values ranging from 0.0033 to 0.38 for other prostate cancer cell lines).
load(url("https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20drug%20sensitivities%20across%20cell%20lines/data_plot_CL.Rdata.txt"))
load(url("https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20drug%20sensitivities%20across%20cell%20lines/correspondance.Rdata.txt"))
listed_nodes <- c("HSPs", "AKT", "TERT", "ERK", "EGFR", "MEK1_2", "PI3K", "AR",
"Caspase8", "cFLAR", "GLUT1", "HIF-1", "MYC_MAX", "p14ARF",
"ROS", "SPOP")
my_cell_lines <- c("BPH-1", "PC-3", "RWPE2-W99", "22RV1", "VCaP", "NCI-H660",
"DU-145", "LNCaP-Clone-FGC", "PWR-1E", "RWPE-1")
options(dplyr.summarise.inform = FALSE)
plot_data <- filter(data_plot_CL, Patient_ID %in% my_cell_lines) %>%
left_join(select(correspondance, Drug_Name, Drug_Target), by = "Drug_Name") %>%
group_by(Patient_ID, Drug_Target, TCGA_DESC) %>%
summarise(Z_SCORE=mean(Z_SCORE), N=n()) %>%
mutate(DR=case_when(
Drug_Target %in% listed_nodes ~ Drug_Target,
TRUE ~ "Other targets"
)) %>%
ungroup %>%
mutate(Patient_ID=if_else(Patient_ID=="LNCaP-Clone-FGC", "LNCap", Patient_ID),
DR=factor(DR, levels=c("AKT", "AR", "EGFR", "ERK", "HSPs", "MEK1_2",
"PI3K", "TERT", "Other targets")))Model-targeting drugs’ sensitivities across prostate cell lines.
GDSC z-score was obtained for all the drugs targeting genes included in the model for all the prostate cell lines in GDSC. Negative values mean that the cell line is more sensitive to the drug. Drugs included in Table 1 were highlighted. ‘Other targets’ are drugs targeting model-related genes that are not part of Table 1.
R code needed to obtain Figure 6.Processed datasets needed are Figure 6—source data 1 and Figure 6—source data 2 are located in the corresponding folder of the repository: here.Processed dataset needed to obtain Figure 6 with Figure 6—source code 1.Processed dataset needed to obtain Figure 6 with Figure 6—source code 1.
chunk: Figure 6
plot_data$Patient_ID2 = factor(plot_data$Patient_ID,
levels=c("LNCap","22RV1","BPH-1","DU-145","PC-3",
"PWR-1E","VCaP"),
labels=c("LNCap","22RV1","BPH-1","DU-145","PC-3",
"PWR-1E","VCaP"))
plot_CL1 <- ggplot(plot_data, aes(y=Z_SCORE)) +
geom_half_boxplot(outlier.shape = NA) +
geom_jitter(aes(x=0.3, color=DR, size=DR, alpha=DR),height = 0, width = 0.2) +
scale_color_manual(values=c("HSPs"="#800000FF", "AKT"="#767676FF",
"ERK"="#FFA319FF","TERT"="#8A9045FF",
"AR"="#155F83FF", "EGFR"="#C16622FF",
"MEK1_2"="#8F3931FF", "PI3K"="#350E20FF",
"Other targets"="grey70")) +
scale_size_manual(values=c("HSPs"=3, "AKT"=3, "ERK"=3, "TERT"=3,"AR"=3,
"EGFR"=3, "MEK1_2"=3, "PI3K"=3,"Other targets"=1)) +
scale_alpha_manual(values=c("HSPs"=1, "AKT"=1, "ERK"=1, "TERT"=1, "AR"=1,
"EGFR"=1, "MEK1_2"=1, "PI3K"=1,"Other targets"=0.5)) +
labs(y="Z-score", color="Target nodes:", size="Target nodes:",
alpha="Target nodes:") +
theme_pubclean() +
theme(legend.position = "top",
legend.title = element_text(face="bold"),
axis.text.x = element_blank(),
axis.title.x = element_blank(),
axis.ticks.x = element_blank(),
legend.key=element_blank()) +
facet_grid(~Patient_ID2)
plot_CL1
::: {figalign="center"}
::: {#fig6}
Overall, the drugs proposed through this analysis suggest the possibility to repurpose drugs that are used in treating other forms of cancer for prostate cancer and open the avenue for further experimental validations based on these suggestions.
Experimental validation of drugs in LNCaP
To validate the model predictions of the candidate drugs, we selected four drugs that target HSPs and PI3K and tested them in LNCaP cell line experiments by using endpoint cell viability measurement assays and real-time cell survival assays using the xCELLigence system (see Materials and methods). The drug selection was a compromise between the drugs identified by our analyses (Table 1) and their effect in diminishing LNCaP’s proliferation (see the previous section). In both assays, drugs that target HSP90AA1 and PI3K/AKT pathway genes retrieved from the model analyses were found to be effective against cell proliferation.
The Hsp90 chaperone is expressed abundantly and plays a crucial role in the correct folding of a wide variety of proteins such as protein kinases and steroid hormone receptors 96Schopf et al.2017. Hsp90 can act as a protector of less stable proteins produced by DNA mutations in cancer cells 7Barrott and Haystead201349Hessenkemper and Baniahmad2013. Currently, Hsp90 inhibitors are in clinical trials for multiple indications in cancer 20Chen et al.202054Iwai et al.201268Le et al.2017. The PI3K/AKT signalling pathway controls many different cellular processes such as cell growth, motility, proliferation, and apoptosis and is frequently altered in different cancer cells 16Carceles-Cordon et al.202098Shorning et al.2020. Many PI3K/AKT inhibitors are in different stages of clinical development, and some of them are approved for clinical use (Table 1).
Notably, Hsp90 (NMS-E973,17-DMAG) and PI3K/AKT pathway (PI-103, Pictilisib) inhibitors showed a dose-dependent activity in the endpoint cell viability assay determined by the fluorescent resazurin after a 48 hr incubation (Figure 7). This dose-dependent activity is more notable in Hsp90 drugs (NMS-E973,17-DMAG) than in PI3K/AKT pathway (Pictilisib) ones and very modest for PI-103.
end1pre <-
read.table(
"https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20experimental%20validation/LNCAPendpoint.txt",
header = TRUE,
sep = "\t",
stringsAsFactors = FALSE
)
end_ids <-
read.table(
"https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20experimental%20validation/LNCAPendpoint_id.txt",
header = TRUE,
sep = "\t",
stringsAsFactors = FALSE
) %>% pivot_longer(-X) %>% mutate(., id = as.integer(gsub("X", "", .$name))) %>%
rename(., drug = X, nM = value) %>% select(-name)
end1 <- full_join(end1pre, end_ids, by = c("drug", "id"))
end1$nM <- as.integer(end1$nM)
end1$count_noise <-
end1$count - (end1 %>% filter(drug == "NoCell") %>% .$count %>% mean())
end1$count_norm <-
end1$count_noise / (end1 %>% filter(drug == "kontroll") %>% .$count_noise %>% mean())
end2 <- end1 %>% filter(!(drug=="NoCell")) %>% mutate(id=factor(id, levels=c(0,5,4,3,2,1))) %>% filter(!(is.na(count)))
end2[is.na(end2$nM),]$id <- 0
end2[is.na(end2$nM),]$nM <- 0.0Cell viability assay determined by the fluorescent resazurin after a 48 hours incubation showed a dose-dependent response to different inhibitors.
(A) Cell viability assay of LNCaP cell line response to 17-DMAG HSP90 inhibitor. (B) Cell viability assay of LNCaP cell line response to PI-103 PI3K/AKT pathway inhibitor. (C) Cell viability assay of LNCaP cell line response to NMS-E973 HSP90 inhibitor. (D) Cell viability assay of LNCaP cell line response to Pictilisib PI3K/AKT pathway inhibitor. Concentrations of drugs were selected to capture their drug-dose response curves. The concentrations for the NMS-E973 are different from the rest as this drug is more potent than the rest (see Materials and methods).
R code needed to obtain Figure 7.Processed datasets needed are Figure 7—source data 1 and 2 and are located in the corresponding folder of the repository: here.Processed dataset needed to obtain Figure 7 with Figure 7—source code 1.Processed dataset needed to obtain with Figure 7—source code 1.
chunk: Figure 7
cols <- c(
"17-DMAG" = pal_uchicago("default")(6)[1],
"NMS-E973" = pal_uchicago("default")(6)[4],
"PI-103" = pal_uchicago("default")(6)[5],
"Pictilisib" = pal_uchicago("default")(6)[6]
)
`17-DMAGa` <-
ggplot() +
theme_bw() +
scale_y_continuous(limits = c(0.25, 1.26)) +
scale_x_discrete(labels=c("0" = "0", "5" = "1", "4" = "5", "3" = "25", "2" = "125", "1" = "625")) +
geom_half_point(data=end2 %>% filter(drug=="17-DMAG" | drug=="kontroll"), aes(x = id, y = count_norm), colour = cols[1], transformation = position_jitter(width = 0.1,height = 0,seed = 4), size = 2) +
geom_half_boxplot(data=end2 %>% filter(drug=="17-DMAG" | drug=="kontroll"), aes(x = id, y = count_norm), fill = cols[1], colour = cols[1], alpha = .3) +
scale_fill_manual(values = cols) + scale_colour_manual(values = cols) +
labs(title = "17-DMAG", x = "Dose (nM)", y = "Normalised cell viability") +
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text =element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank())
`NMS-E973a` <-
ggplot() +
theme_bw() +
scale_y_continuous(limits = c(0.25, 1.26)) +
scale_x_discrete(labels=c("0" = "0", "5" = "2", "4" = "8", "3" = "32", "2" = "128", "1" = "512")) +
geom_half_point(data=end2 %>% filter(drug=="NMS-E973" | drug=="kontroll"), aes(x = id, y = count_norm), colour = cols[2], transformation = position_jitter(width = 0.1,height = 0,seed = 4), size = 2) +
geom_half_boxplot(data=end2 %>% filter(drug=="NMS-E973" | drug=="kontroll"), aes(x = id, y = count_norm), fill = cols[2], colour = cols[2], alpha = .3) +
scale_fill_manual(values = cols) + scale_colour_manual(values = cols) +
labs(title = "NMS-E973", x = "Dose (nM)", y = "Normalised cell viability") +
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text =element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank())
`PI-103a` <-
ggplot() +
theme_bw() +
scale_y_continuous(limits = c(0.25, 1.25)) +
scale_x_discrete(labels=c("0" = "0", "5" = "1", "4" = "5", "3" = "25", "2" = "125", "1" = "625")) +
geom_half_point(data=end2 %>% filter(drug=="PI-103" | drug=="kontroll"), aes(x = id, y = count_norm), colour = cols[3], transformation = position_jitter(width = 0.12,height = 0,seed = 4), size = 2) +
geom_half_boxplot(data=end2 %>% filter(drug=="PI-103" | drug=="kontroll"), aes(x = id, y = count_norm), fill = cols[3], colour = cols[3], alpha = .3) +
scale_fill_manual(values = cols) + scale_colour_manual(values = cols) +
labs(title = "PI-103", x = "Dose (nM)", y = "Normalised cell viability") +
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text =element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank())
Pictilisiba <-
ggplot() +
theme_bw() +
scale_y_continuous(limits = c(0.25, 1.25)) +
scale_x_discrete(labels=c("0" = "0", "5" = "1", "4" = "5", "3" = "25", "2" = "125", "1" = "625")) +
geom_half_point(data=end2 %>% filter(drug=="Pictilisib" | drug=="kontroll"), aes(x = id, y = count_norm), colour = cols[4], transformation = position_jitter(width = 0.12,height = 0,seed = 4), size = 2) +
geom_half_boxplot(data=end2 %>% filter(drug=="Pictilisib" | drug=="kontroll"), aes(x = id, y = count_norm), fill = cols[4], colour = cols[4], alpha = .3) +
scale_fill_manual(values = cols) + scale_colour_manual(values = cols) +
labs(title = "Pictilisib", x = "Dose (nM)", y = "Normalised cell viability") +
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text =element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank())
patchwork = (`17-DMAGa` + `PI-103a`) / (`NMS-E973a` + Pictilisiba)
# Remove titles from subplots
patchwork[[1]][[1]] = patchwork[[1]][[1]] + theme(axis.title.x = element_blank())
patchwork[[1]][[2]] = patchwork[[1]][[2]] + theme(
axis.title.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank()
)
patchwork[[2]][[2]] = patchwork[[2]][[2]] + theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank()
)
patchwork + plot_annotation(tag_levels = "A")
::: {height=5 width=6 figalign="center"}
::: {#fig7}
We studied the real-time response of LNCaP cell viability upon drug addition and saw that the LNCaP cell line is sensitive to Hsp90 and PI3K/AKT pathway inhibitors (Figures 8 and 9, respectively). Both Hsp90 inhibitors tested, 17-DMAG and NMS-E973, reduced the cell viability 12 hr after drug supplementation (Figure 8A for 17-DMAG and Figure 8B for NMS-E973), with 17-DMAG having a stronger effect and in a more clear concentration-dependent manner than NMS-E973 (Appendix 1, Section 8, Appendix 1—figure 40, panels B-D for 17-DMAG and panels F-H for NMS-E973).
b1pre <-
read.table(
"https://github.com/ArnauMontagud/PROFILE_v2/raw/main/Analysis%20of%20experimental%20validation//LNCAP_RT.txt",
header = TRUE,
sep = "\t",
stringsAsFactors = FALSE
)
b1_ids <-
b1pre %>% .[1:2, ] %>% .[-c(1:2)] %>% t() %>% as.data.frame() %>%
rename(., drug = 1, uM = 2) %>% mutate(cell = row.names(.))
b1pre$min <- NA
b1pre$min[-(1:2)] <-
c(as.matrix(read.table(
text = b1pre$Time.Interval[-(1:2)], sep = ":"
)) %*% c(1, 1 / 60, 1 / 3660))
b1 <-
b1pre %>% select(., min, everything(), -contains("Time")) %>% t() %>% as.data.frame()
colnames(b1) <- b1[1, ]
b1 <- b1 %>% .[-1, ]
colnames(b1)[1] <- "drug"
colnames(b1)[2] <- "uM"
b1 %<>% mutate(cell = row.names(.)) %>% select(cell, everything())
b1long <-
b1 %>% pivot_longer(., !(c(drug, uM, cell))) %>% rename(., time = name, CI =
value) %>% as.data.frame()
b1long[which(b1long$drug == "Control"), ]$uM <- 0
b1long$uM <- factor(b1long$uM, levels = c("0", "3.3", "10", "30"))
b1long$time <- as.numeric(b1long$time)
b1long$CI <- as.numeric(b1long$CI)
times <- c("25:41:08", "49:18:09", "74:16:37", "97:17:03")
times_min <-
c(as.matrix(read.table(text = times, sep = ":")) %*% c(1, 1 / 60, 1 / 3660))
times_hour <- c(times_min[1], 48.05246, 72.02869, 96.03388)
times_hour <-
c(times_min[1], times_min[1] + 24, times_min[1] + 48, times_min[1] + 72)
times_hour2 <- times_hour - times_min[1]
b1long <- b1long %>% mutate(time2 = time - times_min[1])
b1long_mean <- b1long %>% filter(grepl("Y.", cell))
b1long_sd <- b1long %>% filter(grepl("SD.", cell))
b2 <- left_join(
b1long %>% filter(grepl("Y.", cell)) %>% rename(mean = CI) %>% .[, -1],
b1long %>% filter(grepl("SD.", cell)) %>% rename(sd = CI) %>% .[, -1],
by = c("drug", "uM", "time", "time2")
)
summary_24 <- left_join(
b1long %>% filter(time > 49 & time < 49.1) %>% filter(grepl("Y.", cell)) %>%
rename(mean = CI) %>% .[, -1],
b1long %>% filter(time > 49 & time < 49.1) %>% filter(grepl("SD.", cell)) %>%
rename(sd = CI) %>% .[, -1],
by = c("drug", "uM", "time", "time2")
)
summary_48 <- left_join(
b1long %>% filter(time > 73 & time < 73.1) %>% filter(grepl("Y.", cell)) %>%
rename(mean = CI) %>% .[, -1],
b1long %>% filter(time > 73 & time < 73.1) %>% filter(grepl("SD.", cell)) %>%
rename(sd = CI) %>% .[, -1],
by = c("drug", "uM", "time", "time2")
)
summary_72 <- left_join(
b1long %>% filter(time > 97 & time < 97.1) %>% filter(grepl("Y.", cell)) %>%
rename(mean = CI) %>% .[, -1],
b1long %>% filter(time > 97 & time < 97.1) %>% filter(grepl("SD.", cell)) %>%
rename(sd = CI) %>% .[, -1],
by = c("drug", "uM", "time", "time2")
)
Hsp90 inhibitors resulted in dose-dependent changes in the LNCaP cell line growth.
(A) Real-time cell electronic sensing (RT-CES) cytotoxicity assay of Hsp90 inhibitor, 17-DMAG, that uses the Cell Index as a measurement of the cell growth rate (see the Materials and methods section). The yellow dotted line represents the 17-DMAG addition. (B) RT-CES cytotoxicity assay of Hsp90 inhibitor, NMS-E973. The yellow dotted line represents the NMS-E973 addition.
Processed dataset to obtain Figures 8 and 9 with Figure 8—source code 1.R code needed to obtain Figures 8 and 9 with Figure 8—source data 1.Processed dataset needed is Figure 8—source data 1 and is located in the corresponding folder of the repository: here.
chunk: Figure 8
pal <- pal_uchicago("default")(9)
# 17-DMAG:
cols <- c(
"0" = pal[2],
"3.3" = "#fa0000",
"10" = "#8f0000",
"30" = "#350000"
)
subdata1 <- b2 %>% filter(drug == "17DMAG" | drug == "Control")
`17-DMAGt_ribbon` <- ggplot() +
theme_bw() +
geom_ribbon(
data = subdata1,
aes(
x = time2,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
fill = uM
),
alpha = 0.2
) +
geom_point(data = subdata1, aes(time2, mean, colour = uM)) +
scale_x_continuous(breaks = seq(-24, 108, 12)) +
scale_colour_manual(name = "Drug dose (uM)", values = cols) +
scale_fill_manual(name = "Drug dose (uM)", values = cols) +
geom_vline(
xintercept = times_hour2,
colour = c(pal[3], pal[7], pal[7], pal[7]),
linetype = "dashed"
) +
ylim(NA, 4) +
labs(title = "", x = "Time (hours)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = c(0.15, 0.6),
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
# NMS-E973:
cols <- c(
"0" = pal[2],
"3.3" = "#c1c960",
"10" = "#6e7337",
"30" = "#292b14"
)
subdata2 <- b2 %>% filter(drug == "NMS-E973" | drug == "Control")
`NMS-E973t_ribbon` <- ggplot() +
theme_bw() +
geom_ribbon(
data = subdata2,
aes(
x = time2,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
fill = uM
),
alpha = 0.2
) +
geom_point(data = subdata2, aes(time2, mean, colour = uM)) +
scale_x_continuous(breaks = seq(-24, 108, 12)) +
scale_colour_manual(name = "Drug dose (uM)", values = cols) +
scale_fill_manual(name = "Drug dose (uM)", values = cols) +
geom_vline(
xintercept = times_hour2,
colour = c(pal[3], pal[7], pal[7], pal[7]),
linetype = "dashed"
) +
ylim(NA, 4) +
labs(title = "", x = "Time (hours)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = c(0.15, 0.6),
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
patchwork_HSP3 = `17-DMAGt_ribbon` / `NMS-E973t_ribbon`
patchwork_HSP3[[1]] = patchwork_HSP3[[1]] + theme(plot.title = element_blank())
patchwork_HSP3[[2]] = patchwork_HSP3[[2]] + theme(plot.title = element_blank())
patchwork_HSP3 + plot_annotation(tag_levels = "A")
::: {height=6 width=7 figalign="center"}
::: {#fig8}
Hsp90 inhibitors resulted in dose-dependent changes in the LNCaP cell line growth.
(A) Real-time cell electronic sensing (RT-CES) cytotoxicity assay of Hsp90 inhibitor, 17-DMAG, that uses the Cell Index as a measurement of the cell growth rate (see the Material and Methods section). The yellow dotted line represents 17-DMAG addition. The brown dotted lines are indicative of the cytotoxicity assay results at 24 hours (B), 48 hours (C) and 72 hours (D) after 17-DMAG addition. (E) RT-CES cytotoxicity assay of Hsp90 inhibitor, NMS-E973. The yellow dotted line represents NMS-E973 addition. The brown dotted lines are indicative of the cytotoxicity assay results at 24 hours (F), 48 hours (G) and 72 hours (H) after NMS-E973 addition.
chunk: Figure S40
# Figure S40: Hsp90 inhibitors
# 17-DMAG:
cols <- c(
"0" = pal[2],
"3.3" = "#fa0000",
"10" = "#8f0000",
"30" = "#350000"
)
`17-DMAG24` <-
ggplot(summary_24 %>% filter(drug == "17DMAG" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "24 hours", x = "Drug dose (uM)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`17-DMAG48` <-
ggplot(summary_48 %>% filter(drug == "17DMAG" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "48 hours", x = "Drug dose (uM)", y = "Cell Index") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`17-DMAG72` <-
ggplot(summary_72 %>% filter(drug == "17DMAG" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "72 hours", x = "Drug dose (uM)", y = "Cell Index") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
# NMS-E973:
cols <- c(
"0" = pal[2],
"3.3" = "#c1c960",
"10" = "#6e7337",
"30" = "#292b14"
)
`NMS-E97324` <-
ggplot(summary_24 %>% filter(drug == "NMS-E973" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "24 hours", x = "Drug dose (uM)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`NMS-E97348` <-
ggplot(summary_48 %>% filter(drug == "NMS-E973" |
drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "48 hours", x = "Drug dose (uM)", y = "Cell Index") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`NMS-E97372` <-
ggplot(summary_72 %>% filter(drug == "NMS-E973" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "72 hours", x = "Drug dose (uM)", y = "Cell Index") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
patchwork_HSP2 = `17-DMAGt_ribbon` /
(`17-DMAG24` + `17-DMAG48` + `17-DMAG72`) /
`NMS-E973t_ribbon` /
(`NMS-E97324` + `NMS-E97348` + `NMS-E97372`)
patchwork_HSP2[[1]] = patchwork_HSP2[[1]] + theme(plot.title = element_blank())
patchwork_HSP2[[2]][[1]] = patchwork_HSP2[[2]][[1]] + theme(plot.title = element_blank())
patchwork_HSP2[[2]][[2]] = patchwork_HSP2[[2]][[2]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_HSP2[[2]][[3]] = patchwork_HSP2[[2]][[3]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_HSP2[[3]] = patchwork_HSP2[[3]] + theme(plot.title = element_blank())
patchwork_HSP2[[4]][[1]] = patchwork_HSP2[[4]][[1]] + theme(plot.title = element_blank())
patchwork_HSP2[[4]][[2]] = patchwork_HSP2[[4]][[2]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_HSP2[[4]][[3]] = patchwork_HSP2[[4]][[3]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_HSP2 + plot_annotation(tag_levels = "A") + plot_layout(heights = c(.7, .3, .7, .3))
::: {height=10 width=7.5 figalign="center"}
::: {#app1fig40}
PI3K/AKT pathway inhibition with different PI3K/AKT inhibitors shows the dose-dependent response in LNCaP cell line growth.
(A) Real-time cell electronic sensing (RT-CES) cytotoxicity assay of PI3K/AKT pathway inhibitor, PI-103, that uses the Cell Index as a measurement of the cell growth rate (see the Materials and methods section). The yellow dotted line represents the PI-103 addition. (B) RT-CES cytotoxicity assay of PI3K/AKT pathway inhibitor, Pictilisib. The yellow dotted line represents the Pictilisib addition.
chunk: Figure 9
# Figure 9: PI3K inhibitors
# PI-103:
cols <- c(
"0" = pal[2],
"3.3" = "#28baff",
"10" = "#176a92",
"30" = "#082736"
)
subdata3 <- b2 %>% filter(drug == "PI-103" | drug == "Control")
`PI-103t_ribbon` <- ggplot() +
theme_bw() +
geom_ribbon(
data = subdata3,
aes(
x = time2,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
fill = uM
),
alpha = 0.2
) +
geom_point(data = subdata3, aes(time2, mean, colour = uM)) +
scale_x_continuous(breaks = seq(-24, 108, 12)) +
scale_colour_manual(name = "Drug dose (uM)", values = cols) +
scale_fill_manual(name = "Drug dose (uM)", values = cols) +
geom_vline(
xintercept = times_hour2,
colour = c(pal[3], pal[7], pal[7], pal[7]),
linetype = "dashed"
) +
ylim(NA, 4) +
labs(title = "", x = "Time (hours)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = c(0.15, 0.6),
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
# Pictilisib:
cols <- c(
"0" = pal[2],
"3.3" = "#ffc641",
"10" = "#cc7125",
"30" = "#4c2a0e"
)
subdata4 <- b2 %>% filter(drug == "Pictilisib" | drug == "Control")
`Pictilisibt_ribbon` <- ggplot() +
theme_bw() +
geom_ribbon(
data = subdata4,
aes(
x = time2,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
fill = uM
),
alpha = 0.2
) +
geom_point(data = subdata4, aes(time2, mean, colour = uM)) +
scale_x_continuous(breaks = seq(-24, 108, 12)) +
scale_colour_manual(name = "Drug dose (uM)", values = cols) +
scale_fill_manual(name = "Drug dose (uM)", values = cols) +
geom_vline(
xintercept = times_hour2,
colour = c(pal[3], pal[7], pal[7], pal[7]),
linetype = "dashed"
) +
ylim(NA, 4) +
labs(title = "", x = "Time (hours)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = c(0.15, 0.6),
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
patchwork_PI3K2 = `PI-103t_ribbon` / `Pictilisibt_ribbon`
patchwork_PI3K2[[1]] = patchwork_PI3K2[[1]] + theme(plot.title = element_blank())
patchwork_PI3K2[[2]] = patchwork_PI3K2[[2]] + theme(plot.title = element_blank())
patchwork_PI3K2 + plot_annotation(tag_levels = "A")
::: {height=6 width=7 figalign="center"}
::: {#fig9}
PI3K/AKT pathway inhibition with different PI3K/AKT inhibitors shows dose-dependent response in LNCaP cell line growth.
(A) Real-time cell electronic sensing (RT-CES) cytotoxicity assay of PI3K/AKT pathway inhibitor, PI-103, that uses the Cell Index as a measurement of the cell growth rate (see the Material and Methods section). The yellow dotted line represents PI-103 addition. The brown dotted lines are indicative of the cytotoxicity assay results at 24 hours (B), 48 hours (C) and 72 hours (D) after PI-103 addition. (E) RT-CES cytotoxicity assay of PI3K/AKT pathway inhibitor, Pictilisib. The yellow dotted line represents Pictilisib addition. The brown dotted lines are indicative of the cytotoxicity assay results at 24 hours (F), 48 hours (G) and 72 hours (H) after Pictilisib addition.
chunk: Figure S41
# Figure S41: PI3K inhibitors
# PI-103:
cols <- c(
"0" = pal[2],
"3.3" = "#28baff",
"10" = "#176a92",
"30" = "#082736"
)
`PI-103t` <-
ggplot(
data = b1long %>% filter(!grepl("SD.", cell)) %>%
filter(drug == "PI-103" | drug == "Control"),
aes(time2, CI, colour = uM)
) +
theme_bw() +
geom_point() +
scale_x_continuous(breaks = seq(-24, 108, 12)) +
scale_colour_manual(name = "Drug dose (uM)", values = cols) +
geom_vline(
xintercept = times_hour2,
colour = c(pal[3], pal[7], pal[7], pal[7]),
linetype = "dashed"
) +
ylim(NA, 4) +
labs(title = "PI-103", x = "Time (hours)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = c(0.15, 0.6),
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`PI-10324` <-
ggplot(summary_24 %>% filter(drug == "PI-103" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "24 hours", x = "Drug dose (uM)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`PI-10348` <-
ggplot(summary_48 %>% filter(drug == "PI-103" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "48 hours", x = "Drug dose (uM)", y = "Cell Index") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`PI-10372` <-
ggplot(summary_72 %>% filter(drug == "PI-103" | drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "72 hours", x = "Drug dose (uM)", y = "Cell Index") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
# Pictilisib:
cols <- c(
"0" = pal[2],
"3.3" = "#ffc641",
"10" = "#cc7125",
"30" = "#4c2a0e"
)
`Pictilisibt` <-
ggplot(
data = b1long %>% filter(!grepl("SD.", cell)) %>% filter(
drug == "Pictilisib" |
drug == "Control"),
aes(time2, CI, colour = uM)
) +
theme_bw() +
geom_point() +
scale_x_continuous(breaks = seq(-24, 108, 12)) +
scale_colour_manual(name = "Drug dose (uM)", values = cols) +
geom_vline(
xintercept = times_hour2,
colour = c(pal[3], pal[7], pal[7], pal[7]),
linetype = "dashed"
) +
ylim(NA, 4) +
labs(title = "Pictilisib", x = "Time (hours)", y = "Cell Index (a.u.)") +
theme(
plot.title = element_text(hjust = 0.5),
legend.position = c(0.15, 0.6),
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`Pictilisib24` <-
ggplot(summary_24 %>% filter(drug == "Pictilisib" |
drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "24 hours", x = "Drug dose (uM)", y = "Cell Index (a.u.)") + theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`Pictilisib48` <-
ggplot(summary_48 %>% filter(drug == "Pictilisib" |
drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "48 hours", x = "Drug dose (uM)", y = "Cell Index") + theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
`Pictilisib72` <-
ggplot(summary_72 %>% filter(drug == "Pictilisib" |
drug == "Control")) +
theme_bw() +
scale_colour_manual(values = cols) +
geom_point(aes(uM, mean, colour = uM))+
geom_errorbar(
aes(
x = uM,
y = mean,
ymin = mean - sd,
ymax = mean + sd,
colour = uM
),
width = .4
) +
scale_y_continuous(limits = c(-1, 4)) +
labs(title = "72 hours", x = "Drug dose (uM)", y = "Cell Index") + theme(
plot.title = element_text(hjust = 0.5),
legend.position = "none",
axis.text = element_text(colour = "black"),
panel.background = element_blank(),
panel.grid.minor = element_blank()
)
patchwork_PI3K = `PI-103t_ribbon` / (`PI-10324` + `PI-10348` + `PI-10372`) / `Pictilisibt_ribbon` / (`Pictilisib24` + `Pictilisib48` + `Pictilisib72`)
patchwork_PI3K[[1]] = patchwork_PI3K[[1]] + theme(plot.title = element_blank())
patchwork_PI3K[[2]][[1]] = patchwork_PI3K[[2]][[1]] + theme(plot.title = element_blank())
patchwork_PI3K[[2]][[2]] = patchwork_PI3K[[2]][[2]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_PI3K[[2]][[3]] = patchwork_PI3K[[2]][[3]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_PI3K[[3]] = patchwork_PI3K[[3]] + theme(plot.title = element_blank())
patchwork_PI3K[[4]][[1]] = patchwork_PI3K[[4]][[1]] + theme(plot.title = element_blank())
patchwork_PI3K[[4]][[2]] = patchwork_PI3K[[4]][[2]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_PI3K[[4]][[3]] = patchwork_PI3K[[4]][[3]] + theme(
plot.title = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
)
patchwork_PI3K + plot_annotation(tag_levels = "A") + plot_layout(heights = c(.7, .3, .7, .3))
::: {height=10 width=7.5 figalign="center"}
::: {#app1fig41}
Likewise, both PI3K/AKT pathway inhibitors tested, Pictilisib and PI-103, reduced the cell viability immediately after drug supplementation (Figure 9A for Pictilisib and Figure 9B for PI-103), in a concentration-dependent manner (Appendix 1, Section 8, Appendix 1—figure 41B-D, for Pictilisib and panels F-H for PI-103). In addition, Hsp90 inhibitors had a more prolonged effect on the cells’ proliferation than PI3K/AKT pathway inhibitors.
Discussion
Clinical assessment of cancers is moving toward more precise, personalised treatments, as the times of one-size-fits-all treatments are no longer appropriate, and patient-tailored models could boost the success rate of these treatments in clinical practice. In this study, we set out to develop a methodology to investigate drug treatments using personalised Boolean models. Our approach consists of building a model that represents the patient-specific disease status and retrieving a list of proposed interventions that affect this disease status, notably by reducing its pro-cancerous behaviours. In this work, we have showcased this methodology by applying it to TCGA prostate cancer patients and to GDSC prostate cancer cell lines, finding patient- and cell line-specific targets and validating selected cell line-specific predicted targets (Figure 1).
First, a prostate cancer Boolean model that encompasses relevant signalling pathways in cancer was constructed based on already published models, experimental data analyses and pathway databases (Figure 2). The influence network and the assignment of logical rules for each node of this network were obtained from known interactions described in the literature (Figure 3). This model describes the regulation of invasion, migration, cell cycle, apoptosis, androgen, and growth factors signalling in prostate cancer (Appendix 1, Section 1).
Second, from this generic Boolean model, we constructed personalised models using the different datasets, that is 488 patients from TCGA and eight cell lines from GDSC. We obtained Gleason score-specific behaviours for TCGA’s patients when studying their Proliferation and Apoptosis scores, observing that high Proliferation scores are higher in high Gleason grades (Figure 4). Thus, the use of these personalised models can help rationalise the relationship of Gleason grading with some of these phenotypes.
Likewise, GDSC data was used with the prostate model to obtain cell line-specific prostate models (Figure 6). These models show differential behaviours, notably in terms of Invasion and Proliferation phenotypes (Appendix 1, Section 5, Appendix 1—figure 21). One of these cell line-specific models, LNCaP, was chosen, and the effects of all its genetic perturbations were thoroughly studied. We studied 32,258 mutants, including single and double mutants, knock-out and over-expressed, and their phenotypes (Appendix 1, Section 6.1, Appendix 1—figures 28 and 29). Thirty-two knock-out perturbations that depleted Proliferation and/or increased Apoptosis were identified, and 16 of them were selected for further analyses (Table 1). The LNCaP-specific model was simulated using different initial conditions that capture different growth media’s specificities, such as RPMI media with and without androgen or epidermal growth factor (Appendix 1, Section 6, Appendix 1—figure 27).
Third, these personalised models were used to simulate the inhibition of druggable genes and proteins, uncovering new treatment’s combination and their synergies. We developed a methodology to simulate drug inhibitions in Boolean models, termed PROFILE_v2, as an extension of previous works 9Béal et al.2019. The LNCaP-specific model was used to obtain simulations with nodes and pairs of nodes corresponding to the genes of interest inhibited with varying strengths. This study allowed us to compile a list of potential targets (Table 1) and to identify potential synergies among genes in the model (Figure 5). Some of the drugs that targeted these genes, such as AKT and TERT, were identified in GDSC as having more sensitivity in LNCaP than in the rest of the prostate cancer cell lines (Figure 6). In addition, drugs that targeted genes included in the model allowed the identification of cell line specificities (Appendix 1, Section 5).
Fourth, we validated the effect of Hsp90 and PI3K/AKT pathway inhibitors on the LNCaP cell line experimentally, finding a concentration-dependent inhibition of the cell line viability as predicted, confirming the role of the drugs targeting these proteins in reducing LNCaP’s proliferation (Figures 7 and 8). Notably, these targets have been studied in other works on prostate cancer 20Chen et al.202068Le et al.2017.
The study presented here enables the study of drug combinations and their synergies. One reason for searching for combinations of drugs is that these have been described for allowing the use of lower doses of each of the two drugs reducing their toxicity 8Bayat Mokhtari et al.2017, evading compensatory mechanisms and combating drug resistances 4Al-Lazikani et al.201263Krzyszczyk et al.2018.
Even if this approach is attractive and promising, it has some limitations. The scope of present work is to test this methodology on a prostate model and infer patient-specific prostate cancer treatments. The method need to be adapted if it were to be expanded to study other cancers by using other models and target lists. The analyses performed with the mathematical model do not aim to predict drug dosages per se but to help in the identification of potential candidates. The patient-specific changes in Proliferation and Apoptosis scores upon mutation are maximal theoretical yields that are used to rank the different potential treatments and should not be used as a direct target for experimental results or clinical trials. Our methodology suggests treatments for individual patients, but the obtained results vary greatly from patient to patient, which is not an uncommon issue of personalised medicine 22Ciccarese et al.201776Molinari et al.2018. This variability is an economic challenge for labs and companies to pursue true patient-specific treatments and also poses challenges in clinical trial designs aimed at validating the model based on the selection of treatments 25Cunanan et al.2017. Nowadays, and because of these constraints, it might be more commercially interesting to target group-specific treatments, which can be more easily related to clinical stages of the disease.
Mathematical modelling of patient profiles helps to classify them in groups with differential characteristics, providing, in essence, a grade-specific treatment. We, therefore, based our analysis on clinical grouping defined by the Gleason grades, but some works have emphasised the difficulty to properly assess them 19Chen and Zhou2016 and, as a result, may not be the perfect predictor for the patient subgrouping in this analysis, even though it is the only available one for these datasets. The lack of subgrouping that stratifies patients adequately may undermine the analysis of our results and could explain the Proliferation and Apoptosis scores of high-grade and low-grade Gleason patients.
Moreover, the behaviours observed in the simulations of the cell line-specific models do not always correspond to what is reported in the literature. The differences between simulation results and biological characteristics could be addressed in further studies by including other pathways, for example, better describing the DNA repair mechanisms, or by tailoring the model with different sets of data, as the data used to personalise these models do not allow for clustering these cell lines according to their different characteristics (Appendix 1, Section 5, Appendix 1—figures 24 and 25). In this sense, another limitation is that we use static data or a snapshot of dynamic data to build dynamic models and to study its stochastic results. Thus, these personalised models would likely improve their performance if they were fitted to dynamic data 94Saez-Rodriguez and Blüthgen2020 or quantitative versions of the models were built, such as ODE-based, that may capture more fine differences among cell lines. As perspectives, we are working on integrating these models in multiscale models to study the effect of the tumour microenvironment 84Leon et al.202185Leon et al.2022, on including information to simulate multiple reagents targeting a single node of the model, on scaling these multiscale models to exascale high-performance computing clusters 78Montagud et al.202195Saxena et al.2021, and on streamlining these studies using workflows in computing clusters to fasten the processing of new, bigger cohorts, as in the PerMedCoE project (https://permedcoe.eu/).
The present work contributes to efforts aimed at using modelling 32Eduati et al.202089Rivas-Barragan et al.202045Gómez Tejeda Zañudo et al.2017 and other computational methods 71Madani Tonekaboni et al.201875Menden et al.2019 for the discovery of novel drug targets and combinatorial strategies. Our study expands the prostate drug catalogue and improves predictions of the impact of these in clinical strategies for prostate cancer by proposing and grading the effectiveness of a set of drugs that could be used off-label or repurposed. The insights gained from this study present the potential of using personalised models to obtain precise, personalised drug treatments for cancer patients.
Materials and methods
Data acquisition
Publicly available data of 489 human prostate cancer patients from TCGA described in 50Hoadley et al.2018 were used in the present work. We gathered mutations, CNA, RNA and clinical data from cBioPortal (https://www.cbioportal.org/study/summary?id=prad_tcga_pan_can_atlas_2018) for all of these samples resulting in 488 with complete omics datasets.
Publicly available data of cell lines used in the present work were obtained from the Genomics of Drug Sensitivity in Cancer database (GDSC) 53Iorio et al.2016. Mutations, CNA and RNA data, as well as cell lines descriptors, were downloaded from (https://www.cancerrxgene.org/downloads). In this work, we have used 3- and 5-stage Gleason grades. Their correspondence is the following: GG Low is GG 1, GG Intermediate is GG 2 and 3, and GG High is GG 4 and 5.
All these data were used to personalise Boolean models using our PROFILE method 9Béal et al.2019.
Prior knowledge network construction
Several sources were used in building this prostate Boolean model and, in particular, the model published by 39Fumiã and Martins2013. This model includes several signalling pathways such as the ones involving receptor tyrosine kinase (RTKs), phosphatidylinositol 3-kinase (PI3K)/AKT, WNT/β-Catenin, transforming growth factor-β (TGF-β)/Smads, cyclins, retinoblastoma protein (Rb), hypoxia-inducible transcription factor (HIF-1), p53 and ataxia-telangiectasia mutated (ATM)/ataxia-telangiectasia and Rad3-related (ATR) protein kinases. The model includes these pathways as well as the substantial cross-talks among them. For a complete description of the process of construction, see Appendix 1, Section 1.
The model also includes several pathways that have a relevant role in our datasets identified by ROMA 74Martignetti et al.2016, a software that uses the first principal component of a PCA analysis to summarise the coexpression of a group of genes in the gene set, identifying significantly overdispersed pathways with a relevant role in a given set of samples. This software was applied to the TCGA transcriptomics data using the gene sets described in the Atlas of Cancer Signaling Networks, ACSN 65Kuperstein et al.2015 (http://www.acsn.curie.fr/) and in the Hallmarks 70Liberzon et al.2015 (Appendix 1, Section 1.1.3, Appendix 1—figure 1) and highlighted the signalling pathways that show high variance across all samples, suggesting candidate pathways and genes. Additionally, OmniPath 111Türei et al.2021 was used to extend the model and complete it, connecting the nodes from Fumiã and Martins and the ones from ROMA analysis. OmniPath is a comprehensive collection of literature-curated human signalling pathways, which includes several databases such as Signor 83Perfetto et al.2016 or Reactome 33Fabregat et al.2016 and that can be queried using pypath, a Python module for molecular networks and pathways analyses.
Fusion genes are frequently found in human prostate cancer and have been identified as a specific subtype marker 15Cancer Genome Atlas Research Network2015. The most frequent is TMPRSS2:ERG, as it involves the transcription factor ERG, which leads to cell-cycle progression. ERG fuses with the AR-regulated TMPRSS2 gene promoter to form an oncogenic fusion gene that is especially common in hormone-refractory prostate cancer, conferring androgen responsiveness to ERG. A literature search reveals that ERG directly regulates EZH2, oncogene c-Myc and many other targets in prostate cancer 64Kunderfranco et al.2010.
We modelled the gene fusion with activation of ERG by the decoupling of ERG in a special node AR_ERG that is only activated by the AR when the fused_event input node is active. In the healthy case, fused_event (that represents TMPRSS2:ERG fusion event) is fixed to 0 or inactive. The occurrence of the gene fusion is represented with the model perturbation where fused_event is fixed to 1. This AR_ERG node is further controlled by tumour suppressor NKX3-1 that accelerates DNA_repair response, and avoids the gene fusion TMPRSS2:ERG. Thus, loss of NKX3-1 favours recruitment to the ERG gene breakpoint of proteins that promote error-prone non-homologous end-joining 12Bowen et al.2015.
The network was further documented using up-to-date literature and was constructed using GINsim 18Chaouiya et al.2012, which allowed us to study its stable states and network properties.
Boolean model construction
We converted the network to a Boolean model by defining a regulatory graph, where each node is associated with discrete levels of activity (0 or 1). Each edge represents a regulatory interaction between the source and target nodes and is labelled with a threshold and a sign (positive or negative). The model is completed by logical rules (or functions), which assign a target value to each node for each regulator level combination 2Abou-Jaoudé et al.201618Chaouiya et al.2012. The regulatory graph was constructed using GINsim software 18Chaouiya et al.2012 and then exported in a format readable by MaBoSS software (see below) in order to perform stochastic simulations on the Boolean model.
The final model has a total of 133 nodes and 449 edges (Supplementary file 1) and includes pathways such as androgen receptor and growth factor signalling, several signalling pathways (Wnt, NFkB, PI3K/AKT, MAPK, mTOR, SHH), cell cycle, epithelial-mesenchymal transition (EMT), Apoptosis, DNA damage, etc. This model has nine inputs (EGF, FGF, TGF beta, Nutrients, Hypoxia, Acidosis, Androgen, TNF alpha, and Carcinogen presence) and six outputs (Proliferation, Apoptosis, Invasion, Migration, (bone) Metastasis, and DNA repair). Note that a node in the network can represent complexes or families of proteins (e.g. AMPK represents the genes PRKAA1, PRKAA2, PRKAB1, PRKAB2, PRKAG1, PRKAG2, PRKAG3). The correspondence can be found in “Montagud2022_interactions_sources.xlsx” and “Montagud2022_nodes_in_pathways.xlsx” in Supplementary file 1.
This model was deposited in the GINsim Database with identifier 252 (http://ginsim.org/model/signalling-prostate-cancer) and in BioModels 72Malik-Sheriff et al.2020 with identifier MODEL2106070001 (https://www.ebi.ac.uk/biomodels/MODEL2106070001). Supplementary file 1 is provided as a zipped folder with the model in several formats: MaBoSS, GINsim, SBML, as well as images of the networks and their annotations. An extensive description of the model construction can be found in the Appendix 1, Section 1.
Stochastic Boolean model simulation
MaBoSS 104Stoll et al.2017103Stoll et al.2012 is a C++ software for stochastically simulating continuous/discrete-time Markov processes defined on the state transition graph (STG) describing the dynamics of a Boolean model (for more details, see 2Abou-Jaoudé et al.2016; 18Chaouiya et al.2012). MaBoSS associates transition rates to each node’s activation and inhibition, enabling it to account for different time scales of the processes described by the model. Probabilities to reach a phenotype (to have value ON) are thus computed by simulating random walks on the probabilistic STG. Since a state in the STG can combine the activation of several phenotypic variables, not all phenotype probabilities are mutually exclusive (like the ones in Appendix 1, Section 6.1, Appendix 1—figure 28). Using MaBoSS, we can study an increase or decrease of a phenotype probability when the model variables are altered (nodes status, initial conditions and transition rates), which may correspond to the effect of particular genetic or environmental perturbation. In the present work, the use of MaBoSS was focused on the readouts of the model, but this can be done for any node of the model.
MaBoSS applies Monte-Carlo kinetic algorithm (i.e. Gillespie algorithm) to the STG to produce time trajectories 104Stoll et al.2017103Stoll et al.2012, so time evolution of probabilities are estimated once a set of initial conditions are defined and a maximum time is set to ensure that the simulations reach asymptotic solutions. Results are analysed in two ways: (1) the trajectories for particular model states (states of nodes) can be interpreted as the evolution of a cell population as a function of time and (2) asymptotic solutions can be represented as pie charts to illustrate the proportions of cells in particular model states. Stochastic simulations with MaBoSS have already been successfully applied to study several Boolean models 13Calzone et al.201023Cohen et al.201587Remy et al.2015. A description of the methods we have used for the simulation of the model can be found in the Appendix 1, Section 2.
Data tailoring the Boolean model
Logical models were tailored to a dataset using PROFILE to obtain personalised models that capture the particularities of a set of patients 9Béal et al.2019 and cell lines 10Béal et al.2021. Proteomics, transcriptomics, mutations and CNA data can be used to modify different variables of the MaBoSS framework, such as node activity status, transition rates and initial conditions. The resulting ensemble of models is a set of personalised variants of the original model that can show great phenotypic differences. Different recipes (use of a given data type to modify a given MaBoSS variable) can be tested to find the combination that better correlates to a given clinical or otherwise descriptive data. In the present case, TCGA patient-specific models were built using mutations, CNA and/or RNA expression data. After studying the effect of these recipes in the clustering of patients according to their Gleason grading (Appendix 1, Section 4.1, Appendix 1—figures 10–14), we chose to use mutations and CNA as discrete data and RNA expression as continuous data.
Likewise, we tried different personalisation recipes to personalise the GDSC prostate cell lines models, but as they had no associated clinical grouping features, we were left with the comparison of the different values for the model’s outputs among the recipes (Appendix 1, Section 5, Appendix 1—figure 23). We used mutation data as discrete data and RNA expression as continuous data as it included the most quantity of data and reproduced the desired results (Appendix 1, Section 5, Appendix 1—figure 23). We decided not to include CNA as discrete data as it forced LNCaP proliferation to be zero by forcing the E2F1 node to be 0 and the SMAD node to be 1 throughout the simulation (for more details, refer to Appendix 1, Section 5).
More on PROFILE’s methodology can be found in its own work 9Béal et al.2019 and at its dedicated GitHub repository (https://github.com/sysbio-curie/PROFILE; 11Béal2022). A description of the methods we have used for the personalisation of the models can be found in the Appendix 1, Section 3. The analysis of the TCGA personalisations and their patient-specific drug treatments can be found in Appendix 1, Section 4. The analysis of the prostate cell lines personalisations can be found in Appendix 1, Section 5, with a special focus on the LNCaP cell line model analysis in Section 6.
High-throughput mutant analysis of Boolean models
MaBoSS allows the study of knock-out or loss-of-function (node forced to 0) and gain-of-function (node forced to 1) mutants as genetic perturbations and of initial conditions as environmental perturbations. Phenotypes’ stabilities against perturbations can be studied and allow to determine driver mutations that promote phenotypic transitions 77Montagud et al.2019.
Genetic interactions were thoroughly studied using our pipeline of computational methods for Boolean modelling of biological networks (available at https://github.com/sysbio-curie/Logical_modelling_pipeline; 80Montagud2022). The LNCaP-specific Boolean model was used to perform single and double knock-out (node forced to 0) and gain-of-function (node forced to 1) mutants for each one of the 133 nodes, resulting in a total of 32,258 models. These were simulated under the same initial conditions, their phenotypic results were collected, and a PCA was applied on the wild type-centred matrix (Appendix 1, Section 6.1, Appendix 1—figures 28 and 29). In addition, we found that the LNCaP model is very robust against perturbations of its logical rules by systematically changing an AND for an OR gate or vice versa in all of its logical rules (Appendix 1, Section 6.2, Appendix 1—figures 30 and 31).
The 488 TCGA patient-specific models were studied in a similar way, but only perturbing 16 nodes from Table 1 shortlisted for their therapeutic target potential (AKT, AR, Caspase8, cFLAR, EGFR, ERK, GLUT1, HIF-1, HSPs, MEK12, MYC_MAX, p14ARF, PI3K, ROS, SPOP, and TERT). Then, the nodes that mostly contributed to a decrease of _Proliferation (Appendix 1, Section 4.2, Appendix 1—figure 19) or an increase in Apoptosis (Appendix 1, Section 4.2, Appendix 1—figure 20) were gathered from the 488 models perturbed.
Additionally, the results of the LNCaP model’s double mutants were used to quantify the level of genetic interactions (epistasis or otherwise) 31Drees et al.2005 between two genetic perturbations (resulting from either the gain-of-function mutation of a gene or from its knock-out or loss-of-function mutation) with respect to wild type phenotypes’ probabilities 14Calzone et al.2015. The method was applied to the LNCaP model studying Proliferation and Apoptosis scores (Appendix 1, Section 7.3.2, Appendix 1—figures 34 and 35).
This genetic interaction study uses the following equation for each gene pair, which is equation 2 in 14Calzone et al.2015:
where and are phenotype fitness values of single gene defects, is the phenotype fitness of the double mutant, and is one of the four functions:
To choose the best definition of , the Pearson correlation coefficient is computed between the fitness values observed in all double mutants and estimated by the null model (more information on 31Drees et al.2005). Regarding the fitness value, to a given phenotype , represents deleterious, beneficial and neutral mutation.
Drug simulations in Boolean models
Logical models can be used to simulate the effect of therapeutic interventions and predict the expected efficacy of candidate drugs on different genetic and environmental backgrounds by using our PROFILEv2 methodology. MaBoSS can perform simulations changing the proportion of activated and inhibited status of a given node. This can be determined in the configuration file of each model (see, for instance, the ‘istate’ section of the CFG files in the Supplementary files 1; 3 and 5). For instance, out of 5,000 trajectories of the Gillespie algorithm, MaBoSS can simulate 70% of them with an activated _AKT and 30% with an inhibited AKT node. The phenotypes’ probabilities for the 5000 trajectories are averaged, and these are considered to be representative of a model with a drug that inhibits 30% of the activity of AKT. The same applies for a combined drug inhibition: a simulation of 50% AKT activity and 50% PI3K will have 50% of them with an activated AKT and 50% with an activated PI3K. Combining them, this will lead to 25% of the trajectories with both AKT and PI3K active, 25% with both nodes inactive, 25% with AKT active and 25% with PI3K active.
In the present work, the LNCaP model has been simulated with different levels of node activity, with 100% of node inhibition (proper knock-out), 80%, 60%, 40%, 20%, and 0% (no inhibition), under four different initial conditions, a nutrient-rich media that simulates RPMI Gibco media with DHT (androgen), with EGF, with both and with none. In terms of the model, the initial conditions are Nutrients is ON and Acidosis, Hypoxia, TGF beta, Carcinogen, and TNF alpha are set to OFF. EGF and Androgen values vary upon simulations. We simulated the inhibition of 17 nodes of interest. These were the 16 nodes from Table 1 with the addition of the fused AR-ERG (Appendix 1, Section 7.3.1, Appendix 1—figures 34 and 35) and their 136 pairwise combinations (Appendix 1, Section 7.3.2, Appendix 1—figures 36 and 37). As we used six different levels of activity for each node, the resulting Appendix 1—figures 36 and 37 comprise a total of 4,998 simulations for each phenotype (136 × 6 x 6 + 17 x 6).
Drug synergies have been studied using Bliss Independence. The Combination Index was calculated with the following equation 37Foucquier and Guedj2015:
where and is the efficiency of the single drug inhibitions and is the inhibition resulting from the double drug simulations. A Combination Index (CI) below 1 represents synergy among drugs (Appendix 1, Section 7.3.2, Appendix 1—figures 36 and 37).
This methodology can be found in its own repository: https://github.com/ArnauMontagud/PROFILE_v2.
Identification of drugs associated with proposed targets
To identify drugs that could act as potential inhibitors of the genes identified with our models (Table 1), we explored the drug-target associations in DrugBank 115Wishart et al.2018. For those genes with multiple drug-target links, only those drugs that are selective and known to have relevance in various forms of cancer are considered here.
In addition to DrugBank searches, we also conducted exhaustive searches in ChEMBL 40Gaulton et al.2017 (http://doi.org/10.6019/CHEMBL.database.23) to suggest potential candidates for genes whose information is not well documented in Drug Bank. From the large number of bioactivities extracted from ChEMBL, we filtered human data and considered only those compounds whose bioactivities fall within a specific threshold (IC50/Kd/ Ki <100 nM).
We performed a target set enrichment analysis using the fgsea method 60Korotkevich et al.2016 from the piano R package 113Väremo et al.2013. We targeted pathway information from the GDSC1 and GDSC2 studies 53Iorio et al.2016 as target sets and performed the enrichment analysis on the normalised drug sensitivity profile of the LNCaP cell line. We normalised drug sensitivity across cell lines in the following way: cells were ranked from most sensitive to least sensitive (using ln(IC50) as the drug sensitivity metrics), and the rank was divided by the number of cell lines tested with the given drug. Thus, the most sensitive cell line has 0, while the most resistant cell line has 1 normalised sensitivity. This rank-based metric made it possible to analyse all drug sensitivities for a given cell line without drug-specific confounding factors, like mean IC50 of a given drug, etc. (Appendix 1, Sections 7.1 and 7.2).
Cell culture method
For the in vitro drug perturbation validations, we used the androgen-sensitive prostate adenocarcinoma cell line LNCaP purchased from American Type Culture Collection (ATCC, Manassas, WV, USA). ATCC found no Mycoplasma contamination and the cell line was identified using STR profiling. Cells were maintained in RPMI-1640 culture media (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) containing 4.5 g/L glucose, 10% foetal bovine serum (FBS, Gibco), 1 X GlutaMAX (Gibco), 1% PenStrep antibiotics (Penicillin G sodium salt, and Streptomycin sulfate salt, Sigma-Aldrich, St. Louis, MI, USA). Cells were maintained in a humidified incubator at 37 °C with 5% CO2 (Sanyo, Osaka, Japan).
Drugs used in the cell culture experiments
We tested two drugs targeted at Hsp90 and two targeted at PI3K complex. 17-DMAG is an Hsp90 inhibitor with an IC50 of 62 nM in a cell-free assay 82Pacey et al.2011. NMS-E973 is an Hsp90 inhibitor with DC50 of <10 nM for Hsp90 binding 36Fogliatto et al.2013. Pictilisib is an inhibitor of PI3K with IC50 of 3.3 nM in cell-free assays 117Zhan et al.2017. PI-103 is a multi-targeted PI3K inhibitor for p110 with IC50 of 2–3 nM in cell-free assays and less potent inhibitor to mTOR/DNA-PK with IC50 of 30 nM 86Raynaud et al.2009. All drugs were obtained from commercial vendors and added to the growth media to have concentrations of 2, 8, 32, 128, and 512 nM for NMS-E973 and 1, 5, 25, 125, and 625 nM for the rest of the drugs in the endpoint cell viability and of 3.3, 10, 30 µM for all the drugs in the RT-CES cytotoxicity assay.
Endpoint cell viability measurements
In vitro toxicity of the selected inhibitors was determined using the viability of LNCaP cells, determined by the fluorescent resazurin (Sigma-Aldrich, Germany) assay as described previously 106Szebeni et al.2017. Briefly, the ∼10,000 LNCaP cells were seeded into 96-well plates (Corning Life Sciences, Tewksbury, MA, USA) in 100 µL RPMI media and incubated overnight. Test compounds were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich, Germany), and cells were treated with an increasing concentration of test compounds: 2, 8, 32, 128, and 512 nM for NMS-E973 and 1, 5, 25, 125, and 625 nM for the rest of the drugs. The highest applied DMSO content of the treated cells was 0.4%. Cell viability was determined after 48 hours of incubation. Resazurin reagent (Sigma–Aldrich, Budapest, Hungary) was added at a final concentration of 25 µg/mL. After 2 hr at 37 °C 5%, CO2 (Sanyo) fluorescence (530 nm excitation/580 nm emission) was recorded on a multimode microplate reader (Cytofluor4000, PerSeptive Biosystems, Framingham, MA, USA). Viability was calculated with relation to blank wells containing media without cells and to wells with untreated cells. Each treatment was repeated in two wells per plate during the experiments, except for the PI-103 treatment with 1 nM in which only one well was used.
In these assays, a deviation of 10–15% for in vitro cellular assays is an acceptable variation as it is a fluorescent assay that detects the cellular metabolic activity of living cells. Thus, in our analyses, we consider changes above 1.00 to be the same value as the controls.
Real-time cell electronic sensing (RT-CES) cytotoxicity assay
A real-time cytotoxicity assay was performed as previously described 81Ozsvári et al.2010. Briefly, RT-CES 96-well E-plate (BioTech Hungary, Budapest, Hungary) was coated with gelatin solution (0.2% in PBS, phosphate buffer saline) for 20 min at 37 °C; then gelatin was washed twice with PBS solution. Growth media (50 µL) was then gently dispensed into each well of the 96-well E-plate for background readings by the RT-CES system prior to the addition of 50 µL of the cell suspension containing 2 × 104 LNCaP cells. Plates were kept at room temperature in a tissue culture hood for 30 min prior to insertion into the RT-CES device in the incubator to allow cells to settle. Cell growth was monitored overnight by measurements of electrical impedance every 15 min. The next day cells were co-treated with different drugs with concentrations of 3.3, 10 and 30 µM. Treated and control wells were dynamically monitored over 72 hr by measurements of electrical impedance every 5 min. Each treatment was repeated in two wells per plate during the experiments, except for the 3.3 µM ones in which only one well was used. Continuous recording of impedance in cells was used as a measurement of the cell growth rate and reflected by the Cell Index value 100Solly et al.2004.
Note that around hour 15, our RT-CES reader had a technical problem caused by a short blackout in our laboratory and the reader detected a minor voltage fluctuation while the uninterruptible power supply (UPS) was switched on. This caused differences that are consistent across all samples and replicates: all wild type and drug reads decrease at that time point, except Pictilisib that slightly increases. For the sake of transparency and as the overall dynamic was not affected, we decided not to remove these readings.
References
- WAbou-Jaoudé
- MChaves
- JLGouzé
- WAbou-Jaoudé
- PTraynard
- PTMonteiro
- JSaez-Rodriguez
- THelikar
- DThieffry
- CChaouiya
- PAdamo
- MRLadomery
- BAl-Lazikani
- UBanerji
- PWorkman
- DCAltieri
- LRLanguino
- JBLian
- JLStein
- ILeav
- AJWijnen
- ZJiang
- GSStein
- ACBabtie
- MPHStumpf
- JJBarrott
- TAJHaystead
- RBayat Mokhtari
- TSHomayouni
- NBaluch
- EMorgatskaya
- SKumar
- BDas
- HYeger
- JBéal
- AMontagud
- PTraynard
- EBarillot
- LCalzone
- JBéal
- LPantolini
- VNoël
- EBarillot
- LCalzone
- JBéal
- CBowen
- TZheng
- EPGelmann
- LCalzone
- LTournier
- SFourquet
- DThieffry
- BZhivotovsky
- EBarillot
- AZinovyev
- LCalzone
- EBarillot
- AZinovyev
- Cancer Genome Atlas Research Network
- MCarceles-Cordon
- WKKelly
- LGomella
- KEKnudsen
- VRodriguez-Bravo
- JDomingo-Domenech
- GCastoria
- LD’Amato
- ACiociola
- PGiovannelli
- TGiraldi
- LSepe
- GPaolella
- MVBarone
- AMigliaccio
- FAuricchio
- CChaouiya
- ANaldi
- DThieffry
- NChen
- QZhou
- WChen
- GLi
- JPeng
- WDai
- QSu
- YHe
- SHCho
- SMPark
- HSLee
- HYLee
- KHCho
- CCiccarese
- FMassari
- RIacovelli
- MFiorentino
- RMontironi
- VDi Nunno
- FGiunchi
- MBrunelli
- GTortora
- DPACohen
- LMartignetti
- SRobine
- EBarillot
- AZinovyev
- LCalzone
- DJColeman
- LGao
- CJKing
- JSchwartzman
- JUrrutia
- ASehrawat
- JTayou
- ABalter
- JBurchard
- KEChiotti
- DSDerrick
- DSun
- ZXia
- LMHeiser
- JJAlumkal
- KMCunanan
- AIasonos
- RShen
- CBBegg
- MGönen
- DCunningham
- ZYou
- MCDaroqui
- PVazquez
- EKier Joffé
- AVBakin
- LIPuricelli
- DDatta
- MAftabuddin
- DKGupta
- SRaha
- PSen
- ODe Wever
- WWestbroek
- AVerloes
- NBloemen
- MBracke
- CGespach
- EBruyneel
- MMareel
- JDorier
- ICrespo
- ANiknejad
- RLiechti
- MEbeling
- IXenarios
- BLDrees
- VThorsson
- GWCarter
- AWRives
- MZRaymond
- IAvila-Campillo
- PShannon
- TGalitski
- FEduati
- PJaaks
- JWappler
- TCramer
- CAMerten
- MJGarnett
- JSaez-Rodriguez
- AFabregat
- KSidiropoulos
- PGarapati
- MGillespie
- KHausmann
- RHaw
- BJassal
- SJupe
- FKorninger
- SMcKay
- LMatthews
- BMay
- MMilacic
- KRothfels
- VShamovsky
- MWebber
- JWeiser
- MWilliams
- GWu
- LStein
- HHermjakob
- PD’Eustachio
- AFauré
- ANaldi
- CChaouiya
- DThieffry
- ÅFlobak
- ABaudot
- ERemy
- LThommesen
- DThieffry
- MKuiper
- ALægreid
- GFogliatto
- LGianellini
- MGBrasca
- ECasale
- DBallinari
- MCiomei
- ADegrassi
- ADe Ponti
- MGermani
- MGuanci
- MPaolucci
- PPolucci
- MRusso
- FSola
- BValsasina
- CVisco
- FZuccotto
- DDonati
- EFelder
- EPesenti
- AGalvani
- SMantegani
- AIsacchi
- JFoucquier
- MGuedj
- JCFrancis
- MKThomsen
- MMTaketo
- ASwain
- HFFumiã
- MLMartins
- AGaulton
- AHersey
- MNowotka
- APBento
- JChambers
- DMendez
- PMutowo
- FAtkinson
- LJBellis
- ECibrián-Uhalte
- MDavies
- NDedman
- AKarlsson
- MPMagariños
- JPOverington
- GPapadatos
- ISmit
- ARLeach
- DTGillespie
- DFGleason
- MTannenbaum
- DFGleason
- AGoldbeter
- JGómez Tejeda Zañudo
- MScaltriti
- RAlbert
- LGrieco
- LCalzone
- IBernard-Pierrot
- FRadvanyi
- BKahn-Perlès
- DThieffry
- SWHayward
- RDahiya
- GRCunha
- JBartek
- NDeshpande
- PNarayan
- THelikar
- JKonvalina
- JHeidel
- JARogers
- WHessenkemper
- ABaniahmad
- KAHoadley
- CYau
- THinoue
- DMWolf
- AJLazar
- EDrill
- RShen
- AMTaylor
- ADCherniack
- VThorsson
- RAkbani
- RBowlby
- CKWong
- MWiznerowicz
- FSanchez-Vega
- AGRobertson
- BGSchneider
- MSLawrence
- HNoushmehr
- TMMalta
- Cancer Genome Atlas Network
- JMStuart
- CCBenz
- PWLaird
- JSHoroszewicz
- SSLeong
- EKawinski
- JPKarr
- HRosenthal
- TMChu
- EAMirand
- GPMurphy
- YHu
- YGu
- HWang
- YHuang
- YMZou
- FIorio
- TAKnijnenburg
- DJVis
- GRBignell
- MPMenden
- MSchubert
- NAben
- EGonçalves
- SBarthorpe
- HLightfoot
- TCokelaer
- PGreninger
- EDyk
- HChang
- HSilva
- HHeyn
- XDeng
- RKEgan
- QLiu
- TMironenko
- XMitropoulos
- LRichardson
- JWang
- TZhang
- SMoran
- SSayols
- MSoleimani
- DTamborero
- NLopez-Bigas
- PRoss-Macdonald
- MEsteller
- NSGray
- DAHaber
- MRStratton
- CHBenes
- LFAWessels
- JSaez-Rodriguez
- UMcDermott
- MJGarnett
- AIwai
- DBourboulia
- MMollapour
- SJensen-Taubman
- SLee
- ACDonnelly
- SYoshida
- NMiyajima
- STsutsumi
- AKSmith
- DSun
- XWu
- BSBlagg
- JBTrepel
- WGStetler-Stevenson
- LNeckers
- BEJohnson
- JWhang-Peng
- SLNaylor
- BZbar
- HBrauch
- ELee
- ASimmons
- ERussell
- MHNam
- AFGazdar
- MEKaighn
- KSNarayan
- YOhnuki
- JFLechner
- LWJones
- SAKauffman
- BNKholodenko
- SSchuster
- JMRohwer
- MCascante
- HVWesterhoff
- SKorenchuk
- JELehr
- LMClean
- YGLee
- SWhitney
- RVessella
- DLLin
- KJPienta
- GKorotkevich
- VSukhov
- ASergushichev
- AKorzybski
- KKrug
- PMertins
- BZhang
- PHornbeck
- RRaju
- RAhmad
- MSzucs
- FMundt
- DForestier
- JJane-Valbuena
- HKeshishian
- MAGillette
- PTamayo
- JPMesirov
- JDJaffe
- SACarr
- DRMani
- PKrzyszczyk
- AAcevedo
- EJDavidoff
- LMTimmins
- IMarrero-Berrios
- MPatel
- CWhite
- CLowe
- JJSherba
- CHartmanshenn
- KMO’Neill
- MLBalter
- ZRFritz
- IPAndroulakis
- RSSchloss
- MLYarmush
- PKunderfranco
- MMello-Grand
- RCangemi
- SPellini
- AMensah
- VAlbertini
- AMalek
- GChiorino
- CVCatapano
- GMCarbone
- IKuperstein
- EBonnet
- HANguyen
- DCohen
- EViara
- LGrieco
- SFourquet
- LCalzone
- CRusso
- MKondratova
- MDutreix
- EBarillot
- AZinovyev
- SLLai
- HBrauch
- TKnutsen
- BEJohnson
- MMNau
- TMitsudomi
- CMTsai
- JWhang-Peng
- BZbar
- FJKaye
- SLé
- JJosse
- FHusson
- BLe
- GLPowers
- YTTam
- NSchumacher
- RLMalinowski
- LSteinke
- GKwon
- PCMarker
- NLe Novère
- ALiberzon
- CBirger
- HThorvaldsdóttir
- MGhandi
- JPMesirov
- PTamayo
- SAMadani Tonekaboni
- LSoltan Ghoraie
- VSKManem
- BHaibe-Kains
- RSMalik-Sheriff
- MGlont
- TVNNguyen
- KTiwari
- MGRoberts
- AXavier
- MTVu
- JMen
- MMaire
- SKananathan
- ELFairbanks
- JPMeyer
- CArankalle
- TMVarusai
- VKnight-Schrijver
- LLi
- CDueñas-Roca
- GDass
- SMKeating
- YMPark
- NBuso
- NRodriguez
- MHucka
- HHermjakob
- CHMarshall
- WFu
- HWang
- ASBaras
- TLLotan
- ESAntonarakis
- LMartignetti
- LCalzone
- EBonnet
- EBarillot
- AZinovyev
- MPMenden
- DWang
- MJMason
- BSzalai
- KCBulusu
- YGuan
- TYu
- JKang
- MJeon
- RWolfinger
- TNguyen
- MZaslavskiy
- AstraZeneca-Sanger Drug Combination DREAM Consortium
- ISJang
- ZGhazoui
- MEAhsen
- RVogel
- ECNeto
- TNorman
- EKYTang
- MJGarnett
- GYDVeroli
- SFawell
- GStolovitzky
- JGuinney
- JRDry
- JSaez-Rodriguez
- CMolinari
- GMarisi
- APassardi
- LMatteucci
- GDe Maio
- PUlivi
- AMontagud
- PTraynard
- LMartignetti
- EBonnet
- EBarillot
- AZinovyev
- LCalzone
- AMontagud
- MLeon
- AValencia
- AMontagud
- AMontagud
- BOzsvári
- LGPuskás
- LINagy
- IKanizsai
- MGyuris
- RMadácsi
- LZFehér
- DGerö
- CSzabó
- SPacey
- RHWilson
- MWalton
- MMEatock
- AHardcastle
- AZetterlund
- H-TArkenau
- JMoreno-Farre
- UBanerji
- BRoels
- HPeachey
- WAherne
- JSBono
- FRaynaud
- PWorkman
- IJudson
- LPerfetto
- LBriganti
- ACalderone
- ACerquone Perpetuini
- MIannuccelli
- FLangone
- LLicata
- MMarinkovic
- AMattioni
- TPavlidou
- DPeluso
- LLPetrilli
- SPirrò
- DPosca
- ESantonico
- ASilvestri
- FSpada
- LCastagnoli
- GCesareni
- MLeon
- AMontagud
- CAkasiadis
- JSchreiber
- TNtiniakou
- AValencia
- MLeon
- AMontagud
- VNoel
- GPradas
- AMeert
- EBarillot
- LCalzone
- AValencia
- FIRaynaud
- SAEccles
- SPatel
- SAlix
- GBox
- IChuckowree
- AFolkes
- SGowan
- ADe Haven Brandon
- FDi Stefano
- AHayes
- ATHenley
- LLensun
- GPergl-Wilson
- ARobson
- NSaghir
- AZhyvoloup
- EMcDonald
- PSheldrake
- SShuttleworth
- MValenti
- NCWan
- PAClarke
- PWorkman
- ERemy
- SRebouissou
- CChaouiya
- AZinovyev
- FRadvanyi
- LCalzone
- GRen
- SBaritaki
- HMarathe
- JFeng
- SPark
- SBeach
- PSBazeley
- ABBeshir
- GFenteany
- RMehra
- SDaignault
- FAl-Mulla
- EKeller
- BBonavida
- ISerna
- KCYeung
- DRivas-Barragan
- SMubeen
- FGuim Bernat
- MHofmann-Apitius
- DDomingo-Fernández
- DRobinson
- EMVan Allen
- Y-MWu
- NSchultz
- RJLonigro
- J-MMosquera
- BMontgomery
- M-ETaplin
- CCPritchard
- GAttard
- HBeltran
- WAbida
- RKBradley
- JVinson
- XCao
- PVats
- LPKunju
- MHussain
- FYFeng
- SATomlins
- KACooney
- DCSmith
- CBrennan
- JSiddiqui
- RMehra
- YChen
- DERathkopf
- MJMorris
- SBSolomon
- JCDurack
- VEReuter
- AGopalan
- JGao
- MLoda
- RTLis
- MBowden
- SPBalk
- GGaviola
- CSougnez
- MGupta
- EYYu
- EAMostaghel
- HHCheng
- HMulcahy
- LDTrue
- SRPlymate
- HDvinge
- RFerraldeschi
- PFlohr
- SMiranda
- ZZafeiriou
- NTunariu
- JMateo
- RPerez-Lopez
- FDemichelis
- BDRobinson
- MSchiffman
- DMNanus
- STTagawa
- ASigaras
- KWEng
- OElemento
- ASboner
- EIHeath
- HIScher
- KJPienta
- PKantoff
- JSBono
- MARubin
- PSNelson
- LAGarraway
- CLSawyers
- AMChinnaiyan
- ARosenblueth
- NWiener
- ASaadatpour
- RAlbert
- JSaez-Rodriguez
- LGAlexopoulos
- JEpperlein
- RSamaga
- DALauffenburger
- SKlamt
- PKSorger
- JSaez-Rodriguez
- NBlüthgen
- GSaxena
- MLeon
- AMontagud
- DVicente Dorca
- AValencia
- ECinquemani
- LPaulevé
- FHSchopf
- MMBiebl
- JBuchner
- LJScott
- BYShorning
- MSDass
- MJSmalley
- HBPearson
- JCSible
- JJTyson
- KSolly
- XWang
- XXu
- BStrulovici
- WZheng
- RMSramkoski
- TGPretlow
- JMGiaconia
- TPPretlow
- SSchwartz
- MSSy
- SRMarengo
- JSRhim
- DZhang
- JWJacobberger
- JSt John
- KPowell
- MKConley-Lacomb
- SRChinni
- GStoll
- EViara
- EBarillot
- LCalzone
- GStoll
- BCaron
- EViara
- ADugourd
- AZinovyev
- ANaldi
- GKroemer
- EBarillot
- LCalzone
- KRStone
- DDMickey
- HWunderli
- GHMickey
- DFPaulson
- GJSzebeni
- ÁBalázs
- IMadarász
- GPócz
- FAyaydin
- IKanizsai
- RFajka-Boja
- RAlföldi
- LHackler
- LGPuskás
- RThomas
- RToth
- HSchiffmann
- CHube-Magg
- FBüscheck
- DHöflmayer
- SWeidemann
- PLebok
- CFraune
- SMinner
- TSchlomm
- GSauter
- CPlass
- YAssenov
- RSimon
- JMeiners
- CGerhäuser
- PTraynard
- AFauré
- FFages
- DThieffry
- DTürei
- TKorcsmáros
- JSaez-Rodriguez
- DTürei
- AValdeolivas
- LGul
- NPalacio-Escat
- MKlein
- OIvanova
- MÖlbei
- AGábor
- FTheis
- DMódos
- TKorcsmáros
- JSaez-Rodriguez
- JJTyson
- TLaomettachit
- PKraikivski
- LVäremo
- JNielsen
- INookaew
- MMWebber
- DBello
- HKKleinman
- DDWartinger
- DEWilliams
- JSRhim
- DSWishart
- YDFeunang
- ACGuo
- EJLo
- AMarcu
- JRGrant
- TSajed
- DJohnson
- CLi
- ZSayeeda
- NAssempour
- IIynkkaran
- YLiu
- AMaciejewski
- NGale
- AWilson
- LChin
- RCummings
- DLe
- APon
- CKnox
- MWilson
- WYang
- MRFreeman
- NKyprianou
- MZhan
- YDeng
- LZhao
- GYan
- FWang
- YTian
- LZhang
- HJiang
- YChen