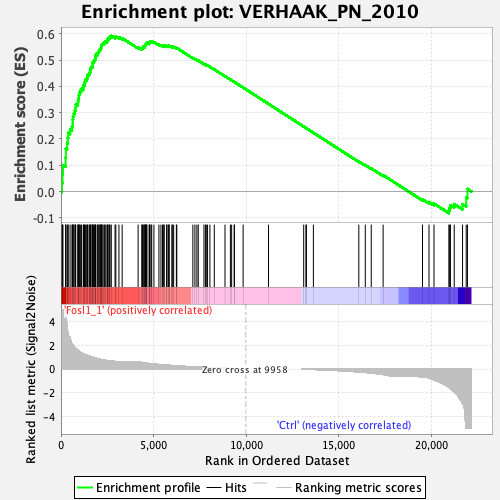
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Fosl1_1 |
| GeneSet | VERHAAK_PN_2010 |
| Enrichment Score (ES) | 0.59211814 |
| Normalized Enrichment Score (NES) | 1.9345015 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SOX10 | SOX10 | 47 | 5.000 | 0.0329 | Yes |
| 2 | LRRTM4 | LRRTM4 | 84 | 5.000 | 0.0663 | Yes |
| 3 | SPTBN2 | SPTBN2 | 104 | 5.000 | 0.1004 | Yes |
| 4 | SCN3A | SCN3A | 248 | 5.000 | 0.1290 | Yes |
| 5 | PAK7 | PAK7 | 261 | 4.993 | 0.1634 | Yes |
| 6 | FXYD6 | FXYD6 | 329 | 3.517 | 0.1850 | Yes |
| 7 | RAB33A | RAB33A | 369 | 2.975 | 0.2040 | Yes |
| 8 | GPR17 | GPR17 | 388 | 2.821 | 0.2230 | Yes |
| 9 | SLC1A1 | SLC1A1 | 491 | 2.601 | 0.2366 | Yes |
| 10 | MAST1 | MAST1 | 592 | 2.093 | 0.2467 | Yes |
| 11 | EPHB1 | EPHB1 | 632 | 2.059 | 0.2594 | Yes |
| 12 | CXXC4 | CXXC4 | 634 | 2.053 | 0.2737 | Yes |
| 13 | IL1RAPL1 | IL1RAPL1 | 654 | 1.998 | 0.2868 | Yes |
| 14 | NLGN3 | NLGN3 | 696 | 1.919 | 0.2984 | Yes |
| 15 | SCG3 | SCG3 | 752 | 1.812 | 0.3086 | Yes |
| 16 | NOL4 | NOL4 | 787 | 1.764 | 0.3194 | Yes |
| 17 | PODXL2 | PODXL2 | 793 | 1.759 | 0.3315 | Yes |
| 18 | VAX2 | VAX2 | 909 | 1.595 | 0.3374 | Yes |
| 19 | DNM3 | DNM3 | 922 | 1.571 | 0.3479 | Yes |
| 20 | YPEL1 | YPEL1 | 953 | 1.532 | 0.3573 | Yes |
| 21 | DUSP26 | DUSP26 | 969 | 1.509 | 0.3672 | Yes |
| 22 | CRB1 | CRB1 | 992 | 1.482 | 0.3765 | Yes |
| 23 | PPM1E | PPM1E | 1046 | 1.420 | 0.3841 | Yes |
| 24 | DGKI | DGKI | 1105 | 1.368 | 0.3910 | Yes |
| 25 | PDE10A | PDE10A | 1203 | 1.288 | 0.3957 | Yes |
| 26 | ABAT | ABAT | 1213 | 1.274 | 0.4042 | Yes |
| 27 | CNTN1 | CNTN1 | 1268 | 1.242 | 0.4104 | Yes |
| 28 | TTYH1 | TTYH1 | 1289 | 1.228 | 0.4181 | Yes |
| 29 | NCALD | NCALD | 1316 | 1.210 | 0.4254 | Yes |
| 30 | SEZ6L | SEZ6L | 1388 | 1.163 | 0.4303 | Yes |
| 31 | MYT1 | MYT1 | 1426 | 1.144 | 0.4367 | Yes |
| 32 | BCAN | BCAN | 1442 | 1.138 | 0.4440 | Yes |
| 33 | HMGB3 | HMGB3 | 1517 | 1.098 | 0.4483 | Yes |
| 34 | GRIA2 | GRIA2 | 1556 | 1.083 | 0.4541 | Yes |
| 35 | DCX | DCX | 1578 | 1.074 | 0.4607 | Yes |
| 36 | TOX3 | TOX3 | 1589 | 1.068 | 0.4677 | Yes |
| 37 | MPPED2 | MPPED2 | 1626 | 1.047 | 0.4734 | Yes |
| 38 | EPB41 | EPB41 | 1694 | 1.010 | 0.4775 | Yes |
| 39 | DLL3 | DLL3 | 1711 | 1.002 | 0.4838 | Yes |
| 40 | MAP2 | MAP2 | 1712 | 1.001 | 0.4908 | Yes |
| 41 | GPM6A | GPM6A | 1751 | 0.979 | 0.4959 | Yes |
| 42 | PCDH11X | PCDH11X | 1804 | 0.958 | 0.5003 | Yes |
| 43 | RALGPS1 | RALGPS1 | 1848 | 0.933 | 0.5048 | Yes |
| 44 | MYO10 | MYO10 | 1853 | 0.931 | 0.5112 | Yes |
| 45 | C1QL1 | C1QL1 | 1865 | 0.923 | 0.5171 | Yes |
| 46 | MAPT | MAPT | 1887 | 0.917 | 0.5226 | Yes |
| 47 | OLIG2 | OLIG2 | 1982 | 0.868 | 0.5244 | Yes |
| 48 | AMOTL2 | AMOTL2 | 1992 | 0.864 | 0.5301 | Yes |
| 49 | DPYSL4 | DPYSL4 | 2045 | 0.842 | 0.5336 | Yes |
| 50 | ATP1A3 | ATP1A3 | 2066 | 0.833 | 0.5385 | Yes |
| 51 | ARHGEF9 | ARHGEF9 | 2122 | 0.810 | 0.5417 | Yes |
| 52 | NRXN1 | NRXN1 | 2154 | 0.799 | 0.5459 | Yes |
| 53 | CSPG5 | CSPG5 | 2171 | 0.792 | 0.5507 | Yes |
| 54 | MARCKSL1 | MARCKSL1 | 2177 | 0.790 | 0.5560 | Yes |
| 55 | FBXO21 | FBXO21 | 2205 | 0.775 | 0.5602 | Yes |
| 56 | DPF1 | DPF1 | 2257 | 0.756 | 0.5632 | Yes |
| 57 | SRGAP3 | SRGAP3 | 2320 | 0.737 | 0.5656 | Yes |
| 58 | CDK5R1 | CDK5R1 | 2357 | 0.734 | 0.5691 | Yes |
| 59 | CLASP2 | CLASP2 | 2418 | 0.727 | 0.5714 | Yes |
| 60 | PAK3 | PAK3 | 2492 | 0.709 | 0.5731 | Yes |
| 61 | REEP1 | REEP1 | 2501 | 0.706 | 0.5777 | Yes |
| 62 | PURG | PURG | 2533 | 0.704 | 0.5812 | Yes |
| 63 | BCL7A | BCL7A | 2589 | 0.695 | 0.5836 | Yes |
| 64 | WASF1 | WASF1 | 2629 | 0.684 | 0.5866 | Yes |
| 65 | RUFY3 | RUFY3 | 2645 | 0.679 | 0.5907 | Yes |
| 66 | SOX11 | SOX11 | 2717 | 0.668 | 0.5921 | Yes |
| 67 | ASCL1 | ASCL1 | 2920 | 0.636 | 0.5874 | No |
| 68 | RAP2A | RAP2A | 2962 | 0.633 | 0.5900 | No |
| 69 | GRID2 | GRID2 | 3130 | 0.631 | 0.5868 | No |
| 70 | RALGPS2 | RALGPS2 | 3304 | 0.602 | 0.5832 | No |
| 71 | SATB1 | SATB1 | 4166 | 0.601 | 0.5483 | No |
| 72 | DBN1 | DBN1 | 4366 | 0.550 | 0.5432 | No |
| 73 | KIF21B | KIF21B | 4393 | 0.542 | 0.5458 | No |
| 74 | CRMP1 | CRMP1 | 4419 | 0.534 | 0.5484 | No |
| 75 | PELI1 | PELI1 | 4446 | 0.528 | 0.5509 | No |
| 76 | GADD45G | GADD45G | 4526 | 0.508 | 0.5509 | No |
| 77 | PLCB4 | PLCB4 | 4533 | 0.505 | 0.5542 | No |
| 78 | MYB | MYB | 4550 | 0.502 | 0.5569 | No |
| 79 | PPM1D | PPM1D | 4569 | 0.498 | 0.5596 | No |
| 80 | MMP16 | MMP16 | 4597 | 0.491 | 0.5618 | No |
| 81 | NRXN2 | NRXN2 | 4628 | 0.483 | 0.5638 | No |
| 82 | MLLT11 | MLLT11 | 4639 | 0.480 | 0.5668 | No |
| 83 | SOX2 | SOX2 | 4761 | 0.460 | 0.5645 | No |
| 84 | CHD7 | CHD7 | 4776 | 0.458 | 0.5671 | No |
| 85 | MARCKS | MARCKS | 4801 | 0.455 | 0.5692 | No |
| 86 | PFN2 | PFN2 | 4863 | 0.443 | 0.5695 | No |
| 87 | SEC61A2 | SEC61A2 | 4901 | 0.436 | 0.5709 | No |
| 88 | TMEFF1 | TMEFF1 | 5020 | 0.418 | 0.5684 | No |
| 89 | MTSS1 | MTSS1 | 5291 | 0.378 | 0.5588 | No |
| 90 | CDKN1B | CDKN1B | 5400 | 0.364 | 0.5565 | No |
| 91 | CSNK1E | CSNK1E | 5484 | 0.353 | 0.5552 | No |
| 92 | GSK3B | GSK3B | 5547 | 0.342 | 0.5548 | No |
| 93 | SOX4 | SOX4 | 5577 | 0.338 | 0.5558 | No |
| 94 | TOP2B | TOP2B | 5705 | 0.321 | 0.5523 | No |
| 95 | SLCO5A1 | SLCO5A1 | 5725 | 0.318 | 0.5537 | No |
| 96 | TTC3 | TTC3 | 5798 | 0.311 | 0.5526 | No |
| 97 | ICK | ICK | 5837 | 0.306 | 0.5530 | No |
| 98 | CASK | CASK | 5839 | 0.306 | 0.5551 | No |
| 99 | ACTR1A | ACTR1A | 5986 | 0.288 | 0.5505 | No |
| 100 | NCAM1 | NCAM1 | 6035 | 0.282 | 0.5503 | No |
| 101 | VEZF1 | VEZF1 | 6086 | 0.277 | 0.5500 | No |
| 102 | MMP15 | MMP15 | 6233 | 0.261 | 0.5452 | No |
| 103 | TMCC1 | TMCC1 | 6256 | 0.258 | 0.5460 | No |
| 104 | CDC25A | CDC25A | 7124 | 0.179 | 0.5079 | No |
| 105 | HDAC2 | HDAC2 | 7235 | 0.170 | 0.5041 | No |
| 106 | BEX1 | BEX1 | 7341 | 0.160 | 0.5005 | No |
| 107 | TSPAN3 | TSPAN3 | 7416 | 0.154 | 0.4982 | No |
| 108 | ZEB2 | ZEB2 | 7724 | 0.136 | 0.4852 | No |
| 109 | ALCAM | ALCAM | 7811 | 0.131 | 0.4822 | No |
| 110 | ATAD5 | ATAD5 | 7846 | 0.130 | 0.4816 | No |
| 111 | PAFAH1B3 | PAFAH1B3 | 7917 | 0.125 | 0.4793 | No |
| 112 | TAF5 | TAF5 | 8045 | 0.116 | 0.4743 | No |
| 113 | MATR3 | MATR3 | 8281 | 0.100 | 0.4644 | No |
| 114 | RAD21 | RAD21 | 8860 | 0.063 | 0.4386 | No |
| 115 | CBX1 | CBX1 | 9151 | 0.043 | 0.4257 | No |
| 116 | FAM110B | FAM110B | 9219 | 0.038 | 0.4230 | No |
| 117 | FHOD3 | FHOD3 | 9356 | 0.028 | 0.4170 | No |
| 118 | STMN4 | STMN4 | 9371 | 0.027 | 0.4165 | No |
| 119 | BCOR | BCOR | 9839 | 0.007 | 0.3954 | No |
| 120 | GABRA3 | GABRA3 | 11211 | 0.000 | 0.3332 | No |
| 121 | STMN1 | STMN1 | 13112 | -0.002 | 0.2470 | No |
| 122 | CKB | CKB | 13233 | -0.005 | 0.2416 | No |
| 123 | LRP6 | LRP6 | 13258 | -0.007 | 0.2405 | No |
| 124 | E2F3 | E2F3 | 13635 | -0.034 | 0.2237 | No |
| 125 | RBPJ | RBPJ | 16089 | -0.243 | 0.1141 | No |
| 126 | TOPBP1 | TOPBP1 | 16439 | -0.288 | 0.1003 | No |
| 127 | P2RX7 | P2RX7 | 16761 | -0.340 | 0.0881 | No |
| 128 | FLRT1 | FLRT1 | 17402 | -0.462 | 0.0623 | No |
| 129 | ERBB3 | ERBB3 | 19530 | -0.663 | -0.0296 | No |
| 130 | CDC7 | CDC7 | 19881 | -0.754 | -0.0402 | No |
| 131 | GSTA4 | GSTA4 | 20151 | -0.923 | -0.0459 | No |
| 132 | GNG4 | GNG4 | 20954 | -1.543 | -0.0715 | No |
| 133 | CLGN | CLGN | 20999 | -1.620 | -0.0621 | No |
| 134 | SORCS3 | SORCS3 | 21038 | -1.662 | -0.0522 | No |
| 135 | MCM10 | MCM10 | 21244 | -1.933 | -0.0480 | No |
| 136 | DPP6 | DPP6 | 21694 | -2.868 | -0.0483 | No |
| 137 | HRASLS | HRASLS | 21892 | -5.000 | -0.0222 | No |
| 138 | FGF9 | FGF9 | 21958 | -5.000 | 0.0099 | No |

