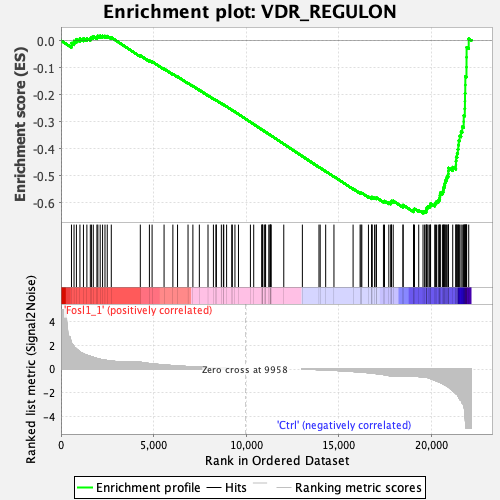
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Ctrl |
| GeneSet | VDR_REGULON |
| Enrichment Score (ES) | -0.6383621 |
| Normalized Enrichment Score (NES) | -2.0052657 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ID4 | ID4 | 566 | 2.195 | -0.0095 | No |
| 2 | PNMA2 | PNMA2 | 704 | 1.902 | -0.0017 | No |
| 3 | PTPRU | PTPRU | 833 | 1.680 | 0.0049 | No |
| 4 | FLI1 | FLI1 | 1023 | 1.450 | 0.0070 | No |
| 5 | ABAT | ABAT | 1213 | 1.274 | 0.0079 | No |
| 6 | PTPRD | PTPRD | 1398 | 1.160 | 0.0081 | No |
| 7 | SPAG1 | SPAG1 | 1584 | 1.070 | 0.0076 | No |
| 8 | SALL2 | SALL2 | 1644 | 1.036 | 0.0125 | No |
| 9 | TPMT | TPMT | 1744 | 0.983 | 0.0153 | No |
| 10 | LLGL2 | LLGL2 | 1941 | 0.888 | 0.0129 | No |
| 11 | TUBB2A | TUBB2A | 1991 | 0.864 | 0.0171 | No |
| 12 | NCAN | NCAN | 2117 | 0.813 | 0.0174 | No |
| 13 | HMBOX1 | HMBOX1 | 2246 | 0.760 | 0.0172 | No |
| 14 | IL2RB | IL2RB | 2376 | 0.731 | 0.0168 | No |
| 15 | IGFBP6 | IGFBP6 | 2499 | 0.706 | 0.0164 | No |
| 16 | SOX11 | SOX11 | 2717 | 0.668 | 0.0115 | No |
| 17 | MAP3K1 | MAP3K1 | 4285 | 0.571 | -0.0554 | No |
| 18 | CHD7 | CHD7 | 4776 | 0.458 | -0.0742 | No |
| 19 | MARK1 | MARK1 | 4921 | 0.433 | -0.0776 | No |
| 20 | NFKBIE | NFKBIE | 5568 | 0.339 | -0.1044 | No |
| 21 | PHLPP1 | PHLPP1 | 6044 | 0.281 | -0.1238 | No |
| 22 | CBR1 | CBR1 | 6296 | 0.254 | -0.1333 | No |
| 23 | ABCA7 | ABCA7 | 6863 | 0.201 | -0.1575 | No |
| 24 | MID2 | MID2 | 7125 | 0.179 | -0.1680 | No |
| 25 | JUP | JUP | 7474 | 0.150 | -0.1827 | No |
| 26 | TRIM24 | TRIM24 | 7942 | 0.123 | -0.2030 | No |
| 27 | ZFR | ZFR | 8234 | 0.103 | -0.2154 | No |
| 28 | DCAF7 | DCAF7 | 8369 | 0.094 | -0.2208 | No |
| 29 | YWHAE | YWHAE | 8395 | 0.093 | -0.2213 | No |
| 30 | C3 | C3 | 8663 | 0.075 | -0.2328 | No |
| 31 | IPPK | IPPK | 8773 | 0.068 | -0.2373 | No |
| 32 | CCND3 | CCND3 | 8793 | 0.067 | -0.2376 | No |
| 33 | SPAG4 | SPAG4 | 8947 | 0.056 | -0.2442 | No |
| 34 | TRAPPC10 | TRAPPC10 | 9225 | 0.038 | -0.2565 | No |
| 35 | PHF7 | PHF7 | 9255 | 0.036 | -0.2575 | No |
| 36 | CPPED1 | CPPED1 | 9397 | 0.026 | -0.2637 | No |
| 37 | ENOX2 | ENOX2 | 9588 | 0.013 | -0.2722 | No |
| 38 | MPZ | MPZ | 10233 | 0.000 | -0.3014 | No |
| 39 | SERPINB7 | SERPINB7 | 10411 | 0.000 | -0.3095 | No |
| 40 | JPH2 | JPH2 | 10848 | 0.000 | -0.3293 | No |
| 41 | SRGN | SRGN | 10870 | 0.000 | -0.3302 | No |
| 42 | DOCK2 | DOCK2 | 10873 | 0.000 | -0.3303 | No |
| 43 | PLEK2 | PLEK2 | 10905 | 0.000 | -0.3317 | No |
| 44 | COL17A1 | COL17A1 | 11004 | 0.000 | -0.3362 | No |
| 45 | GPR39 | GPR39 | 11033 | 0.000 | -0.3374 | No |
| 46 | KYNU | KYNU | 11054 | 0.000 | -0.3383 | No |
| 47 | IL15 | IL15 | 11226 | 0.000 | -0.3461 | No |
| 48 | EXPH5 | EXPH5 | 11290 | 0.000 | -0.3489 | No |
| 49 | CDCP1 | CDCP1 | 11299 | 0.000 | -0.3493 | No |
| 50 | SH3TC1 | SH3TC1 | 11321 | 0.000 | -0.3503 | No |
| 51 | GALNT6 | GALNT6 | 11337 | 0.000 | -0.3509 | No |
| 52 | BANK1 | BANK1 | 11348 | 0.000 | -0.3514 | No |
| 53 | S100A2 | S100A2 | 12035 | 0.000 | -0.3825 | No |
| 54 | RAB27A | RAB27A | 13040 | -0.001 | -0.4281 | No |
| 55 | TRAF3IP1 | TRAF3IP1 | 13937 | -0.055 | -0.4683 | No |
| 56 | TCF12 | TCF12 | 14002 | -0.060 | -0.4708 | No |
| 57 | PLEKHO1 | PLEKHO1 | 14299 | -0.082 | -0.4836 | No |
| 58 | GMDS | GMDS | 14746 | -0.115 | -0.5030 | No |
| 59 | ASPH | ASPH | 15780 | -0.209 | -0.5483 | No |
| 60 | STARD5 | STARD5 | 16159 | -0.251 | -0.5636 | No |
| 61 | TNNC2 | TNNC2 | 16208 | -0.257 | -0.5639 | No |
| 62 | ME1 | ME1 | 16252 | -0.262 | -0.5639 | No |
| 63 | RAD50 | RAD50 | 16610 | -0.314 | -0.5778 | No |
| 64 | IQSEC2 | IQSEC2 | 16780 | -0.345 | -0.5829 | No |
| 65 | ABL2 | ABL2 | 16800 | -0.348 | -0.5812 | No |
| 66 | SVIL | SVIL | 16802 | -0.348 | -0.5787 | No |
| 67 | UBE2Z | UBE2Z | 16941 | -0.371 | -0.5822 | No |
| 68 | LTBR | LTBR | 17023 | -0.385 | -0.5830 | No |
| 69 | CASP4 | CASP4 | 17038 | -0.388 | -0.5808 | No |
| 70 | SMARCD2 | SMARCD2 | 17420 | -0.466 | -0.5946 | No |
| 71 | IL15RA | IL15RA | 17477 | -0.480 | -0.5936 | No |
| 72 | HPCAL1 | HPCAL1 | 17710 | -0.550 | -0.6001 | No |
| 73 | SYNGR2 | SYNGR2 | 17814 | -0.588 | -0.6004 | No |
| 74 | LTBP2 | LTBP2 | 17845 | -0.599 | -0.5974 | No |
| 75 | PDCD1LG2 | PDCD1LG2 | 17855 | -0.599 | -0.5934 | No |
| 76 | PTPRH | PTPRH | 17948 | -0.599 | -0.5931 | No |
| 77 | PRICKLE3 | PRICKLE3 | 18473 | -0.602 | -0.6124 | No |
| 78 | ABLIM3 | ABLIM3 | 18484 | -0.602 | -0.6085 | No |
| 79 | TMEM40 | TMEM40 | 19044 | -0.625 | -0.6292 | No |
| 80 | CDS1 | CDS1 | 19054 | -0.628 | -0.6250 | No |
| 81 | MSX2 | MSX2 | 19097 | -0.630 | -0.6222 | No |
| 82 | ADARB1 | ADARB1 | 19326 | -0.637 | -0.6279 | No |
| 83 | TGM2 | TGM2 | 19558 | -0.671 | -0.6334 | Yes |
| 84 | CHST15 | CHST15 | 19644 | -0.688 | -0.6322 | Yes |
| 85 | STAT6 | STAT6 | 19726 | -0.709 | -0.6306 | Yes |
| 86 | S100A8 | S100A8 | 19732 | -0.709 | -0.6256 | Yes |
| 87 | TNFSF9 | TNFSF9 | 19762 | -0.711 | -0.6217 | Yes |
| 88 | SPINT1 | SPINT1 | 19769 | -0.711 | -0.6167 | Yes |
| 89 | ACSL5 | ACSL5 | 19830 | -0.730 | -0.6141 | Yes |
| 90 | SDC1 | SDC1 | 19916 | -0.776 | -0.6122 | Yes |
| 91 | SLC9A7 | SLC9A7 | 19961 | -0.802 | -0.6083 | Yes |
| 92 | DNASE1L1 | DNASE1L1 | 19967 | -0.805 | -0.6026 | Yes |
| 93 | NAV3 | NAV3 | 20192 | -0.945 | -0.6058 | Yes |
| 94 | CDC42EP1 | CDC42EP1 | 20228 | -0.963 | -0.6003 | Yes |
| 95 | TCIRG1 | TCIRG1 | 20284 | -0.999 | -0.5954 | Yes |
| 96 | IL1R1 | IL1R1 | 20381 | -1.069 | -0.5918 | Yes |
| 97 | G0S2 | G0S2 | 20433 | -1.096 | -0.5861 | Yes |
| 98 | PVR | PVR | 20465 | -1.120 | -0.5792 | Yes |
| 99 | AOX1 | AOX1 | 20473 | -1.128 | -0.5712 | Yes |
| 100 | LAMB3 | LAMB3 | 20486 | -1.139 | -0.5634 | Yes |
| 101 | TNNT1 | TNNT1 | 20605 | -1.219 | -0.5597 | Yes |
| 102 | DENND3 | DENND3 | 20653 | -1.265 | -0.5525 | Yes |
| 103 | GCNT1 | GCNT1 | 20662 | -1.275 | -0.5435 | Yes |
| 104 | HMGA1 | HMGA1 | 20708 | -1.321 | -0.5358 | Yes |
| 105 | FAM114A1 | FAM114A1 | 20738 | -1.339 | -0.5272 | Yes |
| 106 | SLC10A3 | SLC10A3 | 20759 | -1.355 | -0.5182 | Yes |
| 107 | PRKCD | PRKCD | 20816 | -1.410 | -0.5103 | Yes |
| 108 | STEAP3 | STEAP3 | 20857 | -1.442 | -0.5015 | Yes |
| 109 | TNFAIP2 | TNFAIP2 | 20923 | -1.509 | -0.4933 | Yes |
| 110 | ALDH3A1 | ALDH3A1 | 20928 | -1.515 | -0.4823 | Yes |
| 111 | ITPR3 | ITPR3 | 20934 | -1.523 | -0.4713 | Yes |
| 112 | EVI2A | EVI2A | 21163 | -1.831 | -0.4681 | Yes |
| 113 | MME | MME | 21335 | -2.069 | -0.4606 | Yes |
| 114 | GPRC5A | GPRC5A | 21338 | -2.072 | -0.4454 | Yes |
| 115 | PIK3CD | PIK3CD | 21350 | -2.088 | -0.4305 | Yes |
| 116 | FMNL1 | FMNL1 | 21401 | -2.152 | -0.4169 | Yes |
| 117 | AGPAT2 | AGPAT2 | 21438 | -2.245 | -0.4020 | Yes |
| 118 | CYB5R2 | CYB5R2 | 21464 | -2.317 | -0.3861 | Yes |
| 119 | LAMC2 | LAMC2 | 21490 | -2.382 | -0.3696 | Yes |
| 120 | TBX3 | TBX3 | 21538 | -2.532 | -0.3531 | Yes |
| 121 | CXCL1 | CXCL1 | 21612 | -2.603 | -0.3372 | Yes |
| 122 | TRAF1 | TRAF1 | 21676 | -2.868 | -0.3189 | Yes |
| 123 | MYO1D | MYO1D | 21756 | -3.083 | -0.2997 | Yes |
| 124 | STEAP1 | STEAP1 | 21758 | -3.093 | -0.2770 | Yes |
| 125 | LRRC8E | LRRC8E | 21816 | -3.729 | -0.2521 | Yes |
| 126 | PLP2 | PLP2 | 21823 | -3.803 | -0.2243 | Yes |
| 127 | STX11 | STX11 | 21827 | -3.942 | -0.1954 | Yes |
| 128 | FOSL1 | FOSL1 | 21840 | -4.252 | -0.1645 | Yes |
| 129 | TFAP2C | TFAP2C | 21844 | -4.347 | -0.1326 | Yes |
| 130 | ADRB2 | ADRB2 | 21906 | -5.000 | -0.0985 | Yes |
| 131 | PKP3 | PKP3 | 21909 | -5.000 | -0.0617 | Yes |
| 132 | PI15 | PI15 | 21913 | -5.000 | -0.0250 | Yes |
| 133 | SELPLG | SELPLG | 22027 | -5.000 | 0.0068 | Yes |