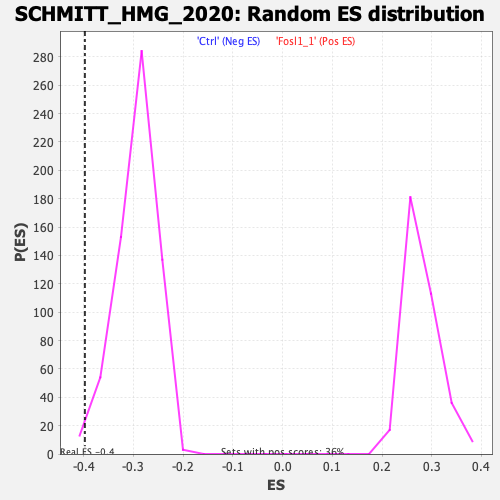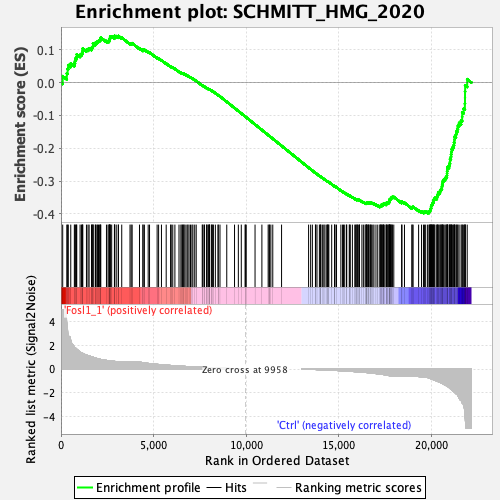
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Ctrl |
| GeneSet | SCHMITT_HMG_2020 |
| Enrichment Score (ES) | -0.3978801 |
| Normalized Enrichment Score (NES) | -1.3514751 |
| Nominal p-value | 0.010869565 |
| FDR q-value | 0.055147376 |
| FWER p-Value | 0.56 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GDA | GDA | 92 | 5.000 | 0.0197 | No |
| 2 | COL11A2 | COL11A2 | 313 | 3.900 | 0.0282 | No |
| 3 | SYN3 | SYN3 | 357 | 3.263 | 0.0419 | No |
| 4 | LIN7A | LIN7A | 393 | 2.756 | 0.0534 | No |
| 5 | FSTL5 | FSTL5 | 527 | 2.410 | 0.0588 | No |
| 6 | NR4A3 | NR4A3 | 721 | 1.858 | 0.0589 | No |
| 7 | EPHA5 | EPHA5 | 755 | 1.812 | 0.0660 | No |
| 8 | ATCAY | ATCAY | 776 | 1.779 | 0.0736 | No |
| 9 | SPTB | SPTB | 839 | 1.674 | 0.0788 | No |
| 10 | AGT | AGT | 850 | 1.666 | 0.0863 | No |
| 11 | GCNT4 | GCNT4 | 1031 | 1.435 | 0.0849 | No |
| 12 | DGKI | DGKI | 1105 | 1.368 | 0.0881 | No |
| 13 | NRCAM | NRCAM | 1147 | 1.335 | 0.0926 | No |
| 14 | ZBTB46 | ZBTB46 | 1169 | 1.321 | 0.0979 | No |
| 15 | RGS3 | RGS3 | 1172 | 1.319 | 0.1042 | No |
| 16 | SYNM | SYNM | 1375 | 1.168 | 0.1005 | No |
| 17 | TCP11L2 | TCP11L2 | 1458 | 1.135 | 0.1022 | No |
| 18 | LIFR | LIFR | 1518 | 1.098 | 0.1047 | No |
| 19 | CHN2 | CHN2 | 1647 | 1.035 | 0.1038 | No |
| 20 | MYCN | MYCN | 1667 | 1.026 | 0.1079 | No |
| 21 | PITPNC1 | PITPNC1 | 1721 | 0.996 | 0.1102 | No |
| 22 | N4BP2L1 | N4BP2L1 | 1728 | 0.992 | 0.1146 | No |
| 23 | KCNK10 | KCNK10 | 1729 | 0.991 | 0.1194 | No |
| 24 | NDRG2 | NDRG2 | 1850 | 0.932 | 0.1183 | No |
| 25 | C1QL1 | C1QL1 | 1865 | 0.923 | 0.1221 | No |
| 26 | COQ8A | COQ8A | 1937 | 0.890 | 0.1231 | No |
| 27 | CISH | CISH | 1967 | 0.875 | 0.1260 | No |
| 28 | EEPD1 | EEPD1 | 2026 | 0.851 | 0.1274 | No |
| 29 | CBS | CBS | 2071 | 0.831 | 0.1293 | No |
| 30 | ATP1B2 | ATP1B2 | 2123 | 0.810 | 0.1309 | No |
| 31 | GLRX | GLRX | 2126 | 0.810 | 0.1347 | No |
| 32 | MEIS3 | MEIS3 | 2150 | 0.801 | 0.1374 | No |
| 33 | ARRDC3 | ARRDC3 | 2456 | 0.723 | 0.1270 | No |
| 34 | IFITM1 | IFITM1 | 2541 | 0.703 | 0.1265 | No |
| 35 | CITED1 | CITED1 | 2597 | 0.692 | 0.1273 | No |
| 36 | FMN2 | FMN2 | 2610 | 0.687 | 0.1300 | No |
| 37 | APOE | APOE | 2626 | 0.685 | 0.1326 | No |
| 38 | PIK3IP1 | PIK3IP1 | 2639 | 0.680 | 0.1353 | No |
| 39 | ABCD2 | ABCD2 | 2655 | 0.676 | 0.1378 | No |
| 40 | DNHD1 | DNHD1 | 2659 | 0.675 | 0.1409 | No |
| 41 | RUNX1T1 | RUNX1T1 | 2738 | 0.661 | 0.1405 | No |
| 42 | VMAC | VMAC | 2886 | 0.643 | 0.1369 | No |
| 43 | ABHD4 | ABHD4 | 2894 | 0.642 | 0.1396 | No |
| 44 | PORCN | PORCN | 2897 | 0.641 | 0.1426 | No |
| 45 | COL14A1 | COL14A1 | 2983 | 0.632 | 0.1417 | No |
| 46 | LAPTM5 | LAPTM5 | 3093 | 0.631 | 0.1398 | No |
| 47 | TRPM8 | TRPM8 | 3102 | 0.631 | 0.1424 | No |
| 48 | FGFBP3 | FGFBP3 | 3281 | 0.608 | 0.1372 | No |
| 49 | SLC22A18 | SLC22A18 | 3728 | 0.602 | 0.1197 | No |
| 50 | BMP5 | BMP5 | 3790 | 0.602 | 0.1198 | No |
| 51 | PTGES | PTGES | 3844 | 0.602 | 0.1202 | No |
| 52 | PPM1K | PPM1K | 4240 | 0.584 | 0.1050 | No |
| 53 | CDKL3 | CDKL3 | 4428 | 0.531 | 0.0990 | No |
| 54 | PLSCR4 | PLSCR4 | 4429 | 0.530 | 0.1015 | No |
| 55 | PFKFB2 | PFKFB2 | 4505 | 0.515 | 0.1005 | No |
| 56 | ZFP28 | ZFP28 | 4710 | 0.469 | 0.0935 | No |
| 57 | CLIP2 | CLIP2 | 4783 | 0.458 | 0.0924 | No |
| 58 | RAB4A | RAB4A | 5193 | 0.391 | 0.0756 | No |
| 59 | MCEE | MCEE | 5273 | 0.380 | 0.0738 | No |
| 60 | YPEL3 | YPEL3 | 5427 | 0.360 | 0.0685 | No |
| 61 | VASH2 | VASH2 | 5683 | 0.324 | 0.0584 | No |
| 62 | MOSPD1 | MOSPD1 | 5920 | 0.296 | 0.0490 | No |
| 63 | RETREG1 | RETREG1 | 5954 | 0.293 | 0.0489 | No |
| 64 | COG6 | COG6 | 6038 | 0.282 | 0.0465 | No |
| 65 | SH3BP5 | SH3BP5 | 6153 | 0.269 | 0.0426 | No |
| 66 | SNHG20 | SNHG20 | 6375 | 0.245 | 0.0336 | No |
| 67 | MYO5B | MYO5B | 6477 | 0.237 | 0.0302 | No |
| 68 | CBX7 | CBX7 | 6533 | 0.232 | 0.0288 | No |
| 69 | OSMR | OSMR | 6568 | 0.229 | 0.0283 | No |
| 70 | PTP4A3 | PTP4A3 | 6629 | 0.222 | 0.0266 | No |
| 71 | TXNIP | TXNIP | 6644 | 0.221 | 0.0270 | No |
| 72 | ACSS2 | ACSS2 | 6705 | 0.216 | 0.0253 | No |
| 73 | FGD4 | FGD4 | 6802 | 0.206 | 0.0219 | No |
| 74 | PROS1 | PROS1 | 6852 | 0.202 | 0.0207 | No |
| 75 | WIPI1 | WIPI1 | 6917 | 0.197 | 0.0187 | No |
| 76 | HABP4 | HABP4 | 6996 | 0.191 | 0.0160 | No |
| 77 | BCAM | BCAM | 7073 | 0.183 | 0.0134 | No |
| 78 | DRAM2 | DRAM2 | 7127 | 0.179 | 0.0119 | No |
| 79 | PIKFYVE | PIKFYVE | 7224 | 0.171 | 0.0083 | No |
| 80 | VGF | VGF | 7304 | 0.163 | 0.0055 | No |
| 81 | SYCP2 | SYCP2 | 7630 | 0.141 | -0.0087 | No |
| 82 | DDR1 | DDR1 | 7658 | 0.140 | -0.0093 | No |
| 83 | RAB3B | RAB3B | 7727 | 0.136 | -0.0117 | No |
| 84 | AOC2 | AOC2 | 7755 | 0.134 | -0.0123 | No |
| 85 | CIR1 | CIR1 | 7867 | 0.128 | -0.0168 | No |
| 86 | MTUS1 | MTUS1 | 7875 | 0.127 | -0.0165 | No |
| 87 | HOTAIRM1 | HOTAIRM1 | 7938 | 0.123 | -0.0187 | No |
| 88 | CLU | CLU | 7980 | 0.121 | -0.0200 | No |
| 89 | ZBTB41 | ZBTB41 | 8012 | 0.119 | -0.0209 | No |
| 90 | YIPF2 | YIPF2 | 8013 | 0.118 | -0.0203 | No |
| 91 | UFM1 | UFM1 | 8034 | 0.117 | -0.0206 | No |
| 92 | YOD1 | YOD1 | 8141 | 0.109 | -0.0250 | No |
| 93 | TBC1D23 | TBC1D23 | 8143 | 0.109 | -0.0245 | No |
| 94 | TMEM87B | TMEM87B | 8194 | 0.105 | -0.0263 | No |
| 95 | RPL37A | RPL37A | 8233 | 0.103 | -0.0275 | No |
| 96 | ENO3 | ENO3 | 8339 | 0.096 | -0.0318 | No |
| 97 | NPC2 | NPC2 | 8485 | 0.088 | -0.0381 | No |
| 98 | SNX10 | SNX10 | 8494 | 0.087 | -0.0380 | No |
| 99 | GLIS3 | GLIS3 | 8589 | 0.080 | -0.0419 | No |
| 100 | UBALD2 | UBALD2 | 8955 | 0.056 | -0.0583 | No |
| 101 | PDE4DIP | PDE4DIP | 9375 | 0.027 | -0.0773 | No |
| 102 | SELENOM | SELENOM | 9581 | 0.013 | -0.0866 | No |
| 103 | PHF24 | PHF24 | 9740 | 0.009 | -0.0938 | No |
| 104 | TMCC3 | TMCC3 | 9950 | 0.001 | -0.1033 | No |
| 105 | KCNA4 | KCNA4 | 9959 | 0.000 | -0.1037 | No |
| 106 | PLPPR4 | PLPPR4 | 10007 | 0.000 | -0.1058 | No |
| 107 | CCNB1IP1 | CCNB1IP1 | 10483 | 0.000 | -0.1275 | No |
| 108 | SLC2A4 | SLC2A4 | 10853 | 0.000 | -0.1444 | No |
| 109 | IL21R | IL21R | 11192 | 0.000 | -0.1598 | No |
| 110 | ITGA11 | ITGA11 | 11245 | 0.000 | -0.1622 | No |
| 111 | APOL6 | APOL6 | 11271 | 0.000 | -0.1633 | No |
| 112 | ICAM1 | ICAM1 | 11338 | 0.000 | -0.1663 | No |
| 113 | HSD3B7 | HSD3B7 | 11441 | 0.000 | -0.1710 | No |
| 114 | SRPX | SRPX | 11916 | 0.000 | -0.1926 | No |
| 115 | CYB561D2 | CYB561D2 | 13376 | -0.015 | -0.2592 | No |
| 116 | STK19 | STK19 | 13487 | -0.023 | -0.2641 | No |
| 117 | TPST1 | TPST1 | 13580 | -0.030 | -0.2681 | No |
| 118 | EIF2AK3 | EIF2AK3 | 13746 | -0.042 | -0.2755 | No |
| 119 | PMM2 | PMM2 | 13761 | -0.044 | -0.2759 | No |
| 120 | ERLEC1 | ERLEC1 | 13830 | -0.048 | -0.2788 | No |
| 121 | GOSR2 | GOSR2 | 13959 | -0.057 | -0.2843 | No |
| 122 | PJA2 | PJA2 | 14004 | -0.061 | -0.2861 | No |
| 123 | POLR3D | POLR3D | 14031 | -0.063 | -0.2869 | No |
| 124 | TMEM231 | TMEM231 | 14038 | -0.063 | -0.2869 | No |
| 125 | FBXO32 | FBXO32 | 14135 | -0.070 | -0.2910 | No |
| 126 | CCNG1 | CCNG1 | 14201 | -0.075 | -0.2936 | No |
| 127 | YPEL2 | YPEL2 | 14261 | -0.079 | -0.2959 | No |
| 128 | PPP2R5A | PPP2R5A | 14357 | -0.085 | -0.2998 | No |
| 129 | HMGCS1 | HMGCS1 | 14392 | -0.088 | -0.3010 | No |
| 130 | EDEM3 | EDEM3 | 14411 | -0.089 | -0.3013 | No |
| 131 | PDXDC1 | PDXDC1 | 14440 | -0.091 | -0.3022 | No |
| 132 | RAB5A | RAB5A | 14471 | -0.093 | -0.3031 | No |
| 133 | ARMCX1 | ARMCX1 | 14624 | -0.107 | -0.3095 | No |
| 134 | HMGXB3 | HMGXB3 | 14782 | -0.119 | -0.3162 | No |
| 135 | GOLGA2 | GOLGA2 | 14789 | -0.119 | -0.3159 | No |
| 136 | ARF4 | ARF4 | 14795 | -0.120 | -0.3155 | No |
| 137 | ACSL6 | ACSL6 | 14885 | -0.125 | -0.3190 | No |
| 138 | PDP1 | PDP1 | 15120 | -0.142 | -0.3290 | No |
| 139 | GOLGA4 | GOLGA4 | 15211 | -0.150 | -0.3324 | No |
| 140 | PLA2G4C | PLA2G4C | 15261 | -0.155 | -0.3339 | No |
| 141 | TSC22D3 | TSC22D3 | 15327 | -0.161 | -0.3361 | No |
| 142 | LBHD1 | LBHD1 | 15423 | -0.172 | -0.3396 | No |
| 143 | HDAC9 | HDAC9 | 15437 | -0.173 | -0.3394 | No |
| 144 | F3 | F3 | 15573 | -0.188 | -0.3446 | No |
| 145 | ZFPL1 | ZFPL1 | 15604 | -0.189 | -0.3451 | No |
| 146 | GPR107 | GPR107 | 15623 | -0.192 | -0.3450 | No |
| 147 | BCL6 | BCL6 | 15742 | -0.205 | -0.3494 | No |
| 148 | PGM2L1 | PGM2L1 | 15874 | -0.220 | -0.3543 | No |
| 149 | USP51 | USP51 | 15905 | -0.224 | -0.3546 | No |
| 150 | FUT1 | FUT1 | 15985 | -0.233 | -0.3571 | No |
| 151 | IRF9 | IRF9 | 15987 | -0.233 | -0.3561 | No |
| 152 | SEC14L1 | SEC14L1 | 16008 | -0.234 | -0.3559 | No |
| 153 | GOLGA5 | GOLGA5 | 16062 | -0.239 | -0.3571 | No |
| 154 | TBC1D15 | TBC1D15 | 16125 | -0.247 | -0.3588 | No |
| 155 | LONRF1 | LONRF1 | 16127 | -0.247 | -0.3577 | No |
| 156 | PLPP5 | PLPP5 | 16256 | -0.262 | -0.3623 | No |
| 157 | MKNK1 | MKNK1 | 16336 | -0.273 | -0.3646 | No |
| 158 | MERTK | MERTK | 16429 | -0.285 | -0.3674 | No |
| 159 | TRIM7 | TRIM7 | 16477 | -0.293 | -0.3681 | No |
| 160 | TAF1D | TAF1D | 16496 | -0.296 | -0.3676 | No |
| 161 | SLC50A1 | SLC50A1 | 16511 | -0.298 | -0.3668 | No |
| 162 | CAMK2D | CAMK2D | 16528 | -0.301 | -0.3661 | No |
| 163 | ACBD3 | ACBD3 | 16544 | -0.304 | -0.3653 | No |
| 164 | MALT1 | MALT1 | 16609 | -0.314 | -0.3667 | No |
| 165 | PDK4 | PDK4 | 16630 | -0.317 | -0.3661 | No |
| 166 | TMEM50B | TMEM50B | 16640 | -0.318 | -0.3650 | No |
| 167 | SPARC | SPARC | 16714 | -0.331 | -0.3668 | No |
| 168 | CRELD1 | CRELD1 | 16737 | -0.335 | -0.3662 | No |
| 169 | GOLGA3 | GOLGA3 | 16764 | -0.342 | -0.3657 | No |
| 170 | GOLGB1 | GOLGB1 | 16845 | -0.354 | -0.3677 | No |
| 171 | SLC26A2 | SLC26A2 | 16907 | -0.364 | -0.3687 | No |
| 172 | LGMN | LGMN | 17016 | -0.384 | -0.3718 | No |
| 173 | S100A10 | S100A10 | 17113 | -0.401 | -0.3743 | No |
| 174 | PODXL | PODXL | 17226 | -0.422 | -0.3774 | No |
| 175 | PIM1 | PIM1 | 17243 | -0.427 | -0.3761 | No |
| 176 | LMAN1 | LMAN1 | 17278 | -0.434 | -0.3756 | No |
| 177 | TMEM120A | TMEM120A | 17299 | -0.439 | -0.3744 | No |
| 178 | SLC30A5 | SLC30A5 | 17304 | -0.440 | -0.3725 | No |
| 179 | SEMA4B | SEMA4B | 17380 | -0.457 | -0.3737 | No |
| 180 | GOLT1B | GOLT1B | 17401 | -0.462 | -0.3724 | No |
| 181 | CIB1 | CIB1 | 17404 | -0.462 | -0.3703 | No |
| 182 | TNFRSF1A | TNFRSF1A | 17424 | -0.466 | -0.3690 | No |
| 183 | NAMPT | NAMPT | 17463 | -0.475 | -0.3684 | No |
| 184 | FZD5 | FZD5 | 17563 | -0.505 | -0.3705 | No |
| 185 | SOCS3 | SOCS3 | 17575 | -0.509 | -0.3686 | No |
| 186 | BLOC1S3 | BLOC1S3 | 17579 | -0.510 | -0.3663 | No |
| 187 | CSF1 | CSF1 | 17655 | -0.533 | -0.3672 | No |
| 188 | SYDE2 | SYDE2 | 17660 | -0.534 | -0.3648 | No |
| 189 | RDH10 | RDH10 | 17719 | -0.554 | -0.3648 | No |
| 190 | DBP | DBP | 17731 | -0.561 | -0.3626 | No |
| 191 | NEDD9 | NEDD9 | 17736 | -0.562 | -0.3601 | No |
| 192 | DUSP5 | DUSP5 | 17740 | -0.564 | -0.3576 | No |
| 193 | ALPK1 | ALPK1 | 17754 | -0.568 | -0.3555 | No |
| 194 | SPTY2D1 | SPTY2D1 | 17800 | -0.584 | -0.3547 | No |
| 195 | SLC2A3 | SLC2A3 | 17819 | -0.591 | -0.3527 | No |
| 196 | EFNA1 | EFNA1 | 17837 | -0.597 | -0.3507 | No |
| 197 | GRIK1 | GRIK1 | 17878 | -0.599 | -0.3496 | No |
| 198 | RPE65 | RPE65 | 17904 | -0.599 | -0.3479 | No |
| 199 | GPR20 | GPR20 | 17979 | -0.599 | -0.3484 | No |
| 200 | LIPG | LIPG | 18401 | -0.601 | -0.3648 | No |
| 201 | GPX3 | GPX3 | 18421 | -0.602 | -0.3628 | No |
| 202 | IDI1 | IDI1 | 18556 | -0.602 | -0.3660 | No |
| 203 | IFITM2 | IFITM2 | 18954 | -0.603 | -0.3813 | No |
| 204 | SC5D | SC5D | 18974 | -0.609 | -0.3792 | No |
| 205 | C4A | C4A | 19023 | -0.624 | -0.3784 | No |
| 206 | PKIG | PKIG | 19320 | -0.632 | -0.3889 | No |
| 207 | AKNA | AKNA | 19478 | -0.651 | -0.3930 | No |
| 208 | SGK1 | SGK1 | 19582 | -0.680 | -0.3944 | No |
| 209 | CDHR1 | CDHR1 | 19642 | -0.688 | -0.3939 | No |
| 210 | ARID5A | ARID5A | 19701 | -0.706 | -0.3931 | No |
| 211 | ZFP36 | ZFP36 | 19806 | -0.715 | -0.3945 | Yes |
| 212 | VMP1 | VMP1 | 19880 | -0.754 | -0.3942 | Yes |
| 213 | PRICKLE2 | PRICKLE2 | 19905 | -0.770 | -0.3916 | Yes |
| 214 | MAN2A1 | MAN2A1 | 19935 | -0.788 | -0.3892 | Yes |
| 215 | CRTAP | CRTAP | 19959 | -0.801 | -0.3864 | Yes |
| 216 | DNAJB5 | DNAJB5 | 19978 | -0.812 | -0.3834 | Yes |
| 217 | MSMO1 | MSMO1 | 19995 | -0.820 | -0.3802 | Yes |
| 218 | TNC | TNC | 19997 | -0.820 | -0.3763 | Yes |
| 219 | KLF9 | KLF9 | 20018 | -0.836 | -0.3732 | Yes |
| 220 | A2M | A2M | 20053 | -0.858 | -0.3707 | Yes |
| 221 | PDGFRB | PDGFRB | 20067 | -0.868 | -0.3671 | Yes |
| 222 | MVP | MVP | 20100 | -0.891 | -0.3643 | Yes |
| 223 | INSIG1 | INSIG1 | 20135 | -0.914 | -0.3615 | Yes |
| 224 | PMP22 | PMP22 | 20136 | -0.915 | -0.3572 | Yes |
| 225 | LYST | LYST | 20163 | -0.926 | -0.3539 | Yes |
| 226 | GBP2 | GBP2 | 20202 | -0.952 | -0.3511 | Yes |
| 227 | TRIQK | TRIQK | 20303 | -1.011 | -0.3509 | Yes |
| 228 | ISG20 | ISG20 | 20319 | -1.025 | -0.3466 | Yes |
| 229 | CDKN1A | CDKN1A | 20334 | -1.035 | -0.3423 | Yes |
| 230 | PDP2 | PDP2 | 20371 | -1.060 | -0.3389 | Yes |
| 231 | GADD45A | GADD45A | 20380 | -1.069 | -0.3342 | Yes |
| 232 | MAFF | MAFF | 20468 | -1.123 | -0.3328 | Yes |
| 233 | CD44 | CD44 | 20484 | -1.138 | -0.3281 | Yes |
| 234 | KLF6 | KLF6 | 20529 | -1.164 | -0.3245 | Yes |
| 235 | COL6A1 | COL6A1 | 20578 | -1.210 | -0.3209 | Yes |
| 236 | EPHX1 | EPHX1 | 20582 | -1.211 | -0.3153 | Yes |
| 237 | TMEM97 | TMEM97 | 20594 | -1.214 | -0.3100 | Yes |
| 238 | AHNAK | AHNAK | 20623 | -1.238 | -0.3054 | Yes |
| 239 | TLCD2 | TLCD2 | 20631 | -1.243 | -0.2997 | Yes |
| 240 | EMP1 | EMP1 | 20675 | -1.290 | -0.2955 | Yes |
| 241 | FAM114A1 | FAM114A1 | 20738 | -1.339 | -0.2920 | Yes |
| 242 | CRYAB | CRYAB | 20793 | -1.381 | -0.2879 | Yes |
| 243 | NUAK2 | NUAK2 | 20847 | -1.435 | -0.2834 | Yes |
| 244 | RCN3 | RCN3 | 20855 | -1.441 | -0.2769 | Yes |
| 245 | TAGLN2 | TAGLN2 | 20860 | -1.446 | -0.2702 | Yes |
| 246 | EML2 | EML2 | 20870 | -1.453 | -0.2636 | Yes |
| 247 | ITGA10 | ITGA10 | 20881 | -1.463 | -0.2571 | Yes |
| 248 | FABP3 | FABP3 | 20964 | -1.551 | -0.2534 | Yes |
| 249 | TTC39B | TTC39B | 20983 | -1.594 | -0.2467 | Yes |
| 250 | LDLR | LDLR | 21016 | -1.633 | -0.2403 | Yes |
| 251 | TIMP1 | TIMP1 | 21019 | -1.640 | -0.2326 | Yes |
| 252 | MAP3K6 | MAP3K6 | 21063 | -1.705 | -0.2264 | Yes |
| 253 | JAK3 | JAK3 | 21074 | -1.720 | -0.2186 | Yes |
| 254 | MATN2 | MATN2 | 21088 | -1.736 | -0.2110 | Yes |
| 255 | CCDC113 | CCDC113 | 21104 | -1.758 | -0.2032 | Yes |
| 256 | NUPR1 | NUPR1 | 21158 | -1.828 | -0.1969 | Yes |
| 257 | TMEM173 | TMEM173 | 21215 | -1.892 | -0.1905 | Yes |
| 258 | KDELR3 | KDELR3 | 21234 | -1.921 | -0.1821 | Yes |
| 259 | DTX3L | DTX3L | 21258 | -1.965 | -0.1738 | Yes |
| 260 | THBS2 | THBS2 | 21263 | -1.969 | -0.1646 | Yes |
| 261 | AHNAK2 | AHNAK2 | 21336 | -2.071 | -0.1580 | Yes |
| 262 | TENT5C | TENT5C | 21356 | -2.119 | -0.1487 | Yes |
| 263 | COL6A2 | COL6A2 | 21407 | -2.158 | -0.1407 | Yes |
| 264 | SERPINE1 | SERPINE1 | 21444 | -2.262 | -0.1316 | Yes |
| 265 | JADE2 | JADE2 | 21506 | -2.439 | -0.1227 | Yes |
| 266 | RGS16 | RGS16 | 21614 | -2.607 | -0.1151 | Yes |
| 267 | JAG2 | JAG2 | 21672 | -2.868 | -0.1041 | Yes |
| 268 | TRAF1 | TRAF1 | 21676 | -2.868 | -0.0905 | Yes |
| 269 | BST2 | BST2 | 21754 | -3.070 | -0.0794 | Yes |
| 270 | PTCHD1 | PTCHD1 | 21820 | -3.762 | -0.0644 | Yes |
| 271 | LHX9 | LHX9 | 21826 | -3.827 | -0.0463 | Yes |
| 272 | ATF3 | ATF3 | 21828 | -3.948 | -0.0275 | Yes |
| 273 | ATG9B | ATG9B | 21834 | -4.077 | -0.0083 | Yes |
| 274 | IL13RA1 | IL13RA1 | 21945 | -5.000 | 0.0105 | Yes |