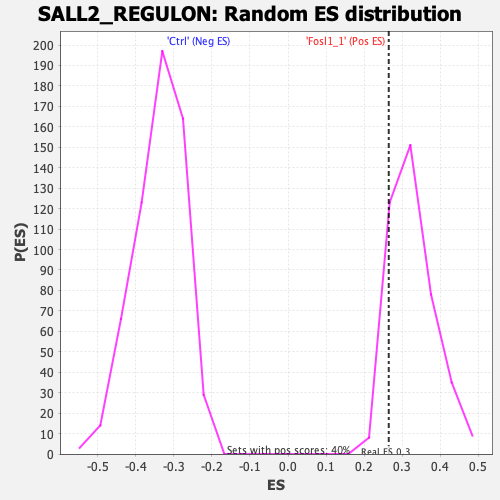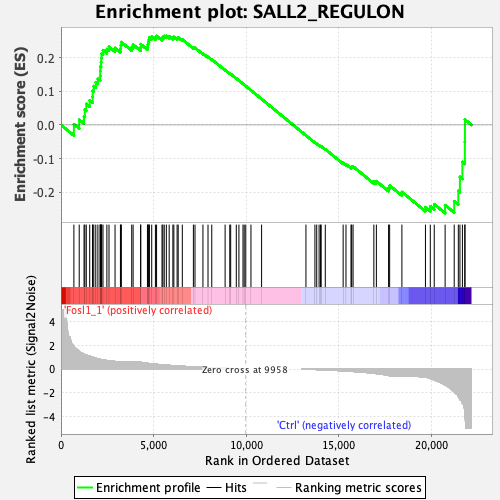
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Fosl1_1 |
| GeneSet | SALL2_REGULON |
| Enrichment Score (ES) | 0.26491842 |
| Normalized Enrichment Score (NES) | 0.8103148 |
| Nominal p-value | 0.90841585 |
| FDR q-value | 0.8761875 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NLGN3 | NLGN3 | 696 | 1.919 | 0.0022 | Yes |
| 2 | APBA2 | APBA2 | 984 | 1.493 | 0.0155 | Yes |
| 3 | ENPP1 | ENPP1 | 1245 | 1.250 | 0.0257 | Yes |
| 4 | TTYH1 | TTYH1 | 1289 | 1.228 | 0.0453 | Yes |
| 5 | RNFT2 | RNFT2 | 1366 | 1.173 | 0.0625 | Yes |
| 6 | SLC22A17 | SLC22A17 | 1551 | 1.085 | 0.0732 | Yes |
| 7 | PCP4 | PCP4 | 1707 | 1.003 | 0.0838 | Yes |
| 8 | MAP2 | MAP2 | 1712 | 1.001 | 0.1012 | Yes |
| 9 | RASSF2 | RASSF2 | 1773 | 0.970 | 0.1156 | Yes |
| 10 | ACSBG1 | ACSBG1 | 1879 | 0.919 | 0.1269 | Yes |
| 11 | ALDH6A1 | ALDH6A1 | 1993 | 0.864 | 0.1370 | Yes |
| 12 | NCAN | NCAN | 2117 | 0.813 | 0.1457 | Yes |
| 13 | ATP1B2 | ATP1B2 | 2123 | 0.810 | 0.1597 | Yes |
| 14 | CTNND2 | CTNND2 | 2127 | 0.809 | 0.1738 | Yes |
| 15 | NOTCH3 | NOTCH3 | 2163 | 0.795 | 0.1862 | Yes |
| 16 | MARCKSL1 | MARCKSL1 | 2177 | 0.790 | 0.1995 | Yes |
| 17 | MSI1 | MSI1 | 2197 | 0.783 | 0.2124 | Yes |
| 18 | EFNB3 | EFNB3 | 2268 | 0.752 | 0.2224 | Yes |
| 19 | FZD3 | FZD3 | 2478 | 0.715 | 0.2255 | Yes |
| 20 | NOVA1 | NOVA1 | 2593 | 0.694 | 0.2326 | Yes |
| 21 | ASCL1 | ASCL1 | 2920 | 0.636 | 0.2290 | Yes |
| 22 | JAM2 | JAM2 | 3211 | 0.627 | 0.2269 | Yes |
| 23 | NTAN1 | NTAN1 | 3233 | 0.620 | 0.2368 | Yes |
| 24 | EVL | EVL | 3258 | 0.613 | 0.2465 | Yes |
| 25 | PAQR6 | PAQR6 | 3821 | 0.602 | 0.2316 | Yes |
| 26 | GAL3ST4 | GAL3ST4 | 3896 | 0.602 | 0.2388 | Yes |
| 27 | FLRT3 | FLRT3 | 4301 | 0.566 | 0.2305 | Yes |
| 28 | ARNT2 | ARNT2 | 4308 | 0.565 | 0.2402 | Yes |
| 29 | DTX3 | DTX3 | 4670 | 0.475 | 0.2322 | Yes |
| 30 | ZMYM2 | ZMYM2 | 4694 | 0.472 | 0.2394 | Yes |
| 31 | MT3 | MT3 | 4714 | 0.469 | 0.2468 | Yes |
| 32 | ZHX2 | ZHX2 | 4750 | 0.463 | 0.2533 | Yes |
| 33 | CHD7 | CHD7 | 4776 | 0.458 | 0.2603 | Yes |
| 34 | SEC61A2 | SEC61A2 | 4901 | 0.436 | 0.2623 | Yes |
| 35 | KIF1B | KIF1B | 5101 | 0.405 | 0.2604 | Yes |
| 36 | NPAS3 | NPAS3 | 5163 | 0.397 | 0.2646 | Yes |
| 37 | RAB8B | RAB8B | 5457 | 0.356 | 0.2576 | Yes |
| 38 | MYLK | MYLK | 5503 | 0.349 | 0.2617 | Yes |
| 39 | SOX4 | SOX4 | 5577 | 0.338 | 0.2644 | Yes |
| 40 | PCM1 | PCM1 | 5691 | 0.323 | 0.2649 | Yes |
| 41 | AGRN | AGRN | 5844 | 0.306 | 0.2634 | No |
| 42 | PHLPP1 | PHLPP1 | 6044 | 0.281 | 0.2593 | No |
| 43 | ZFP30 | ZFP30 | 6097 | 0.276 | 0.2618 | No |
| 44 | CHD8 | CHD8 | 6273 | 0.257 | 0.2584 | No |
| 45 | RIN2 | RIN2 | 6331 | 0.251 | 0.2603 | No |
| 46 | DENND5A | DENND5A | 6554 | 0.230 | 0.2543 | No |
| 47 | SEMA6A | SEMA6A | 7152 | 0.177 | 0.2303 | No |
| 48 | GLRX2 | GLRX2 | 7239 | 0.170 | 0.2294 | No |
| 49 | STARD7 | STARD7 | 7668 | 0.139 | 0.2125 | No |
| 50 | TRIM24 | TRIM24 | 7942 | 0.123 | 0.2023 | No |
| 51 | TRAPPC3 | TRAPPC3 | 8145 | 0.108 | 0.1951 | No |
| 52 | CDC42BPB | CDC42BPB | 8866 | 0.062 | 0.1636 | No |
| 53 | CEP68 | CEP68 | 9118 | 0.045 | 0.1530 | No |
| 54 | DICER1 | DICER1 | 9167 | 0.042 | 0.1515 | No |
| 55 | DIP2C | DIP2C | 9470 | 0.021 | 0.1382 | No |
| 56 | POMT2 | POMT2 | 9612 | 0.012 | 0.1321 | No |
| 57 | TARBP1 | TARBP1 | 9832 | 0.008 | 0.1223 | No |
| 58 | KIF2A | KIF2A | 9897 | 0.004 | 0.1195 | No |
| 59 | ADAMTS3 | ADAMTS3 | 9982 | 0.000 | 0.1157 | No |
| 60 | KL | KL | 10262 | 0.000 | 0.1030 | No |
| 61 | IL17RB | IL17RB | 10837 | 0.000 | 0.0770 | No |
| 62 | CKB | CKB | 13233 | -0.005 | -0.0313 | No |
| 63 | MED8 | MED8 | 13718 | -0.040 | -0.0525 | No |
| 64 | SGSM2 | SGSM2 | 13815 | -0.047 | -0.0560 | No |
| 65 | AGPAT5 | AGPAT5 | 13944 | -0.055 | -0.0608 | No |
| 66 | YKT6 | YKT6 | 14022 | -0.062 | -0.0632 | No |
| 67 | DHTKD1 | DHTKD1 | 14055 | -0.065 | -0.0635 | No |
| 68 | MEF2D | MEF2D | 14278 | -0.080 | -0.0722 | No |
| 69 | PLXNA2 | PLXNA2 | 15243 | -0.153 | -0.1131 | No |
| 70 | LLGL1 | LLGL1 | 15396 | -0.168 | -0.1171 | No |
| 71 | COPB2 | COPB2 | 15667 | -0.196 | -0.1258 | No |
| 72 | BAG5 | BAG5 | 15704 | -0.200 | -0.1240 | No |
| 73 | PDCD6IP | PDCD6IP | 15779 | -0.209 | -0.1236 | No |
| 74 | VASH1 | VASH1 | 16902 | -0.364 | -0.1680 | No |
| 75 | CASP4 | CASP4 | 17038 | -0.388 | -0.1673 | No |
| 76 | OSTF1 | OSTF1 | 17697 | -0.544 | -0.1876 | No |
| 77 | TANC2 | TANC2 | 17755 | -0.568 | -0.1802 | No |
| 78 | WNT11 | WNT11 | 18417 | -0.602 | -0.1995 | No |
| 79 | ARPC1B | ARPC1B | 19689 | -0.703 | -0.2447 | No |
| 80 | SNX7 | SNX7 | 19951 | -0.797 | -0.2425 | No |
| 81 | MICAL2 | MICAL2 | 20164 | -0.927 | -0.2358 | No |
| 82 | LMNA | LMNA | 20752 | -1.350 | -0.2387 | No |
| 83 | PTGS2 | PTGS2 | 21243 | -1.932 | -0.2269 | No |
| 84 | STX3 | STX3 | 21465 | -2.317 | -0.1962 | No |
| 85 | FXYD5 | FXYD5 | 21552 | -2.588 | -0.1546 | No |
| 86 | TBC1D8B | TBC1D8B | 21686 | -2.868 | -0.1102 | No |
| 87 | PROCR | PROCR | 21818 | -3.729 | -0.0507 | No |
| 88 | PLP2 | PLP2 | 21823 | -3.803 | 0.0160 | No |