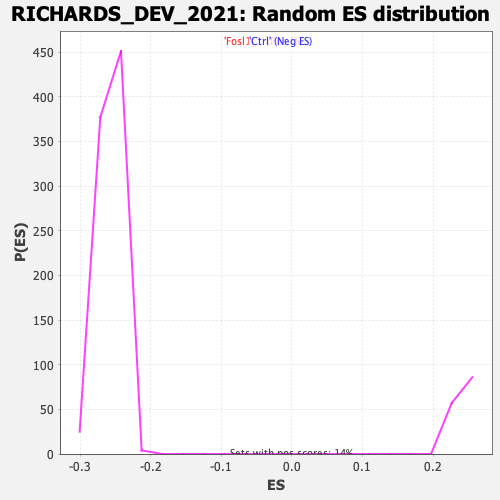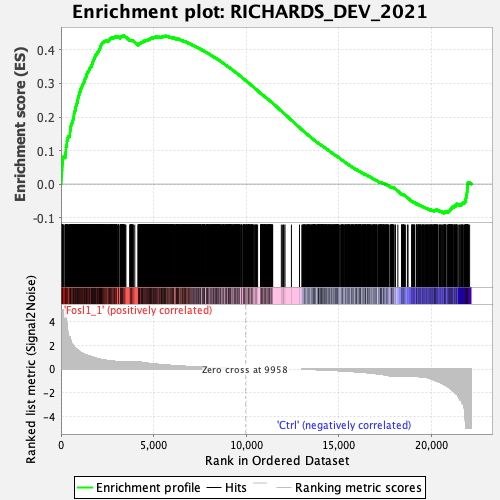
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Fosl1_1 |
| GeneSet | RICHARDS_DEV_2021 |
| Enrichment Score (ES) | 0.44201216 |
| Normalized Enrichment Score (NES) | 1.8099186 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.79842E-4 |
| FWER p-Value | 0.003 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CACNA1E | CACNA1E | 4 | 5.000 | 0.0028 | Yes |
| 2 | TTC9B | TTC9B | 8 | 5.000 | 0.0057 | Yes |
| 3 | SPIRE2 | SPIRE2 | 10 | 5.000 | 0.0087 | Yes |
| 4 | EYA2 | EYA2 | 13 | 5.000 | 0.0117 | Yes |
| 5 | CACNA1G | CACNA1G | 16 | 5.000 | 0.0146 | Yes |
| 6 | CHGA | CHGA | 17 | 5.000 | 0.0177 | Yes |
| 7 | FGF17 | FGF17 | 21 | 5.000 | 0.0205 | Yes |
| 8 | DCT | DCT | 22 | 5.000 | 0.0236 | Yes |
| 9 | FAIM2 | FAIM2 | 27 | 5.000 | 0.0264 | Yes |
| 10 | PCSK2 | PCSK2 | 37 | 5.000 | 0.0290 | Yes |
| 11 | HPCA | HPCA | 42 | 5.000 | 0.0319 | Yes |
| 12 | CPLX1 | CPLX1 | 48 | 5.000 | 0.0347 | Yes |
| 13 | DGKB | DGKB | 50 | 5.000 | 0.0376 | Yes |
| 14 | KCNJ9 | KCNJ9 | 55 | 5.000 | 0.0405 | Yes |
| 15 | FMO2 | FMO2 | 58 | 5.000 | 0.0434 | Yes |
| 16 | SLC24A4 | SLC24A4 | 62 | 5.000 | 0.0463 | Yes |
| 17 | SVOP | SVOP | 63 | 5.000 | 0.0494 | Yes |
| 18 | RTN4R | RTN4R | 66 | 5.000 | 0.0523 | Yes |
| 19 | CCR10 | CCR10 | 67 | 5.000 | 0.0554 | Yes |
| 20 | ENTPD1 | ENTPD1 | 77 | 5.000 | 0.0579 | Yes |
| 21 | STKLD1 | STKLD1 | 79 | 5.000 | 0.0609 | Yes |
| 22 | NANOS2 | NANOS2 | 82 | 5.000 | 0.0639 | Yes |
| 23 | RYR3 | RYR3 | 89 | 5.000 | 0.0666 | Yes |
| 24 | HIST1H4A | HIST1H4A | 93 | 5.000 | 0.0695 | Yes |
| 25 | SRRM4 | SRRM4 | 97 | 5.000 | 0.0724 | Yes |
| 26 | SCN1A | SCN1A | 99 | 5.000 | 0.0754 | Yes |
| 27 | SNORD35B | SNORD35B | 100 | 5.000 | 0.0784 | Yes |
| 28 | GPR179 | GPR179 | 109 | 5.000 | 0.0811 | Yes |
| 29 | SOX1 | SOX1 | 168 | 5.000 | 0.0812 | Yes |
| 30 | KCNQ2 | KCNQ2 | 233 | 5.000 | 0.0810 | Yes |
| 31 | LY6H | LY6H | 234 | 5.000 | 0.0840 | Yes |
| 32 | ACTL6B | ACTL6B | 238 | 5.000 | 0.0869 | Yes |
| 33 | SCN3A | SCN3A | 248 | 5.000 | 0.0895 | Yes |
| 34 | FOXB1 | FOXB1 | 251 | 5.000 | 0.0925 | Yes |
| 35 | FAM181A | FAM181A | 257 | 5.000 | 0.0953 | Yes |
| 36 | PAK7 | PAK7 | 261 | 4.993 | 0.0981 | Yes |
| 37 | CELF6 | CELF6 | 266 | 4.828 | 0.1009 | Yes |
| 38 | SLITRK3 | SLITRK3 | 267 | 4.828 | 0.1038 | Yes |
| 39 | SYT2 | SYT2 | 270 | 4.819 | 0.1066 | Yes |
| 40 | NEB | NEB | 271 | 4.819 | 0.1096 | Yes |
| 41 | EPHA10 | EPHA10 | 283 | 4.736 | 0.1119 | Yes |
| 42 | GSG1L | GSG1L | 302 | 4.122 | 0.1135 | Yes |
| 43 | BRSK2 | BRSK2 | 303 | 4.093 | 0.1160 | Yes |
| 44 | SPAG8 | SPAG8 | 315 | 3.871 | 0.1178 | Yes |
| 45 | IGDCC3 | IGDCC3 | 316 | 3.861 | 0.1201 | Yes |
| 46 | DSCAML1 | DSCAML1 | 320 | 3.766 | 0.1223 | Yes |
| 47 | HEY2 | HEY2 | 321 | 3.765 | 0.1246 | Yes |
| 48 | CNKSR2 | CNKSR2 | 322 | 3.745 | 0.1269 | Yes |
| 49 | FXYD6 | FXYD6 | 329 | 3.517 | 0.1287 | Yes |
| 50 | COL9A1 | COL9A1 | 332 | 3.493 | 0.1307 | Yes |
| 51 | ADGRB1 | ADGRB1 | 351 | 3.369 | 0.1319 | Yes |
| 52 | KANSL2 | KANSL2 | 360 | 3.200 | 0.1334 | Yes |
| 53 | NEUROD1 | NEUROD1 | 367 | 3.016 | 0.1349 | Yes |
| 54 | RAB33A | RAB33A | 369 | 2.975 | 0.1367 | Yes |
| 55 | CBLN1 | CBLN1 | 370 | 2.975 | 0.1385 | Yes |
| 56 | CRYL1 | CRYL1 | 379 | 2.894 | 0.1399 | Yes |
| 57 | GPR17 | GPR17 | 388 | 2.821 | 0.1412 | Yes |
| 58 | GRIA3 | GRIA3 | 391 | 2.756 | 0.1428 | Yes |
| 59 | ADARB2 | ADARB2 | 444 | 2.725 | 0.1418 | Yes |
| 60 | SCRT2 | SCRT2 | 446 | 2.725 | 0.1434 | Yes |
| 61 | FBXO41 | FBXO41 | 475 | 2.723 | 0.1436 | Yes |
| 62 | LRP2BP | LRP2BP | 477 | 2.723 | 0.1452 | Yes |
| 63 | DTX1 | DTX1 | 481 | 2.689 | 0.1467 | Yes |
| 64 | ROBO2 | ROBO2 | 483 | 2.671 | 0.1483 | Yes |
| 65 | PIANP | PIANP | 484 | 2.661 | 0.1499 | Yes |
| 66 | PRKCZ | PRKCZ | 486 | 2.653 | 0.1515 | Yes |
| 67 | SMPD3 | SMPD3 | 487 | 2.648 | 0.1531 | Yes |
| 68 | ZCCHC18 | ZCCHC18 | 489 | 2.623 | 0.1546 | Yes |
| 69 | SLC1A1 | SLC1A1 | 491 | 2.601 | 0.1562 | Yes |
| 70 | BSN | BSN | 492 | 2.600 | 0.1578 | Yes |
| 71 | FAM222A | FAM222A | 496 | 2.584 | 0.1592 | Yes |
| 72 | TNN | TNN | 497 | 2.579 | 0.1608 | Yes |
| 73 | ASPDH | ASPDH | 512 | 2.517 | 0.1616 | Yes |
| 74 | SLC24A3 | SLC24A3 | 513 | 2.517 | 0.1631 | Yes |
| 75 | CHRNB2 | CHRNB2 | 515 | 2.512 | 0.1646 | Yes |
| 76 | SHD | SHD | 517 | 2.470 | 0.1660 | Yes |
| 77 | KCNJ10 | KCNJ10 | 518 | 2.470 | 0.1675 | Yes |
| 78 | SLC35D3 | SLC35D3 | 526 | 2.418 | 0.1687 | Yes |
| 79 | GJC3 | GJC3 | 528 | 2.399 | 0.1701 | Yes |
| 80 | PACRG | PACRG | 529 | 2.394 | 0.1715 | Yes |
| 81 | STK32C | STK32C | 535 | 2.369 | 0.1727 | Yes |
| 82 | CSRNP3 | CSRNP3 | 538 | 2.349 | 0.1740 | Yes |
| 83 | USP2 | USP2 | 544 | 2.314 | 0.1752 | Yes |
| 84 | SLITRK1 | SLITRK1 | 549 | 2.263 | 0.1764 | Yes |
| 85 | WNT7A | WNT7A | 560 | 2.223 | 0.1772 | Yes |
| 86 | ID4 | ID4 | 566 | 2.195 | 0.1783 | Yes |
| 87 | WNT3 | WNT3 | 569 | 2.194 | 0.1795 | Yes |
| 88 | KIF5A | KIF5A | 581 | 2.140 | 0.1803 | Yes |
| 89 | RAB3C | RAB3C | 589 | 2.105 | 0.1812 | Yes |
| 90 | MAST1 | MAST1 | 592 | 2.093 | 0.1824 | Yes |
| 91 | CSMD1 | CSMD1 | 602 | 2.093 | 0.1832 | Yes |
| 92 | MPC1 | MPC1 | 609 | 2.077 | 0.1842 | Yes |
| 93 | GPR137C | GPR137C | 627 | 2.076 | 0.1846 | Yes |
| 94 | SHC2 | SHC2 | 631 | 2.059 | 0.1857 | Yes |
| 95 | EPHB1 | EPHB1 | 632 | 2.059 | 0.1869 | Yes |
| 96 | CXXC4 | CXXC4 | 634 | 2.053 | 0.1881 | Yes |
| 97 | DSCAM | DSCAM | 640 | 2.035 | 0.1891 | Yes |
| 98 | ARHGAP33 | ARHGAP33 | 645 | 2.020 | 0.1901 | Yes |
| 99 | RGCC | RGCC | 648 | 2.015 | 0.1913 | Yes |
| 100 | ENKUR | ENKUR | 653 | 1.998 | 0.1923 | Yes |
| 101 | MCF2L | MCF2L | 659 | 1.990 | 0.1932 | Yes |
| 102 | SRPK3 | SRPK3 | 665 | 1.976 | 0.1942 | Yes |
| 103 | ARHGEF26 | ARHGEF26 | 671 | 1.954 | 0.1951 | Yes |
| 104 | IL17D | IL17D | 685 | 1.937 | 0.1956 | Yes |
| 105 | UNC80 | UNC80 | 686 | 1.937 | 0.1968 | Yes |
| 106 | SERINC4 | SERINC4 | 687 | 1.929 | 0.1980 | Yes |
| 107 | TMEM255A | TMEM255A | 689 | 1.929 | 0.1991 | Yes |
| 108 | MAPK8IP2 | MAPK8IP2 | 691 | 1.924 | 0.2002 | Yes |
| 109 | RLBP1 | RLBP1 | 692 | 1.924 | 0.2014 | Yes |
| 110 | SCRT1 | SCRT1 | 693 | 1.921 | 0.2026 | Yes |
| 111 | NLGN3 | NLGN3 | 696 | 1.919 | 0.2037 | Yes |
| 112 | COL27A1 | COL27A1 | 697 | 1.913 | 0.2048 | Yes |
| 113 | RFX4 | RFX4 | 699 | 1.913 | 0.2059 | Yes |
| 114 | CELF3 | CELF3 | 700 | 1.913 | 0.2071 | Yes |
| 115 | SLC6A1 | SLC6A1 | 701 | 1.910 | 0.2083 | Yes |
| 116 | PNMA2 | PNMA2 | 704 | 1.902 | 0.2093 | Yes |
| 117 | OPRL1 | OPRL1 | 707 | 1.888 | 0.2104 | Yes |
| 118 | PSD2 | PSD2 | 713 | 1.879 | 0.2113 | Yes |
| 119 | TUBB4A | TUBB4A | 716 | 1.869 | 0.2123 | Yes |
| 120 | FGD3 | FGD3 | 727 | 1.843 | 0.2129 | Yes |
| 121 | LGI3 | LGI3 | 733 | 1.824 | 0.2138 | Yes |
| 122 | DBX2 | DBX2 | 737 | 1.823 | 0.2147 | Yes |
| 123 | ST6GAL2 | ST6GAL2 | 738 | 1.822 | 0.2158 | Yes |
| 124 | B3GALT1 | B3GALT1 | 740 | 1.819 | 0.2169 | Yes |
| 125 | SCG3 | SCG3 | 752 | 1.812 | 0.2174 | Yes |
| 126 | GABBR2 | GABBR2 | 759 | 1.807 | 0.2182 | Yes |
| 127 | ELOVL2 | ELOVL2 | 760 | 1.806 | 0.2193 | Yes |
| 128 | GBX2 | GBX2 | 766 | 1.799 | 0.2202 | Yes |
| 129 | GRIK4 | GRIK4 | 775 | 1.781 | 0.2209 | Yes |
| 130 | ATCAY | ATCAY | 776 | 1.779 | 0.2219 | Yes |
| 131 | ROBO1 | ROBO1 | 777 | 1.778 | 0.2230 | Yes |
| 132 | WNK2 | WNK2 | 779 | 1.774 | 0.2240 | Yes |
| 133 | ATP10B | ATP10B | 781 | 1.772 | 0.2251 | Yes |
| 134 | AP3B2 | AP3B2 | 785 | 1.768 | 0.2260 | Yes |
| 135 | NOL4 | NOL4 | 787 | 1.764 | 0.2270 | Yes |
| 136 | PIP5K1B | PIP5K1B | 791 | 1.762 | 0.2279 | Yes |
| 137 | PODXL2 | PODXL2 | 793 | 1.759 | 0.2290 | Yes |
| 138 | CNIH2 | CNIH2 | 797 | 1.750 | 0.2299 | Yes |
| 139 | FGFR3 | FGFR3 | 800 | 1.743 | 0.2308 | Yes |
| 140 | CELSR3 | CELSR3 | 806 | 1.732 | 0.2316 | Yes |
| 141 | HLF | HLF | 816 | 1.709 | 0.2322 | Yes |
| 142 | KCNJ16 | KCNJ16 | 819 | 1.704 | 0.2332 | Yes |
| 143 | ELAVL4 | ELAVL4 | 823 | 1.701 | 0.2340 | Yes |
| 144 | SARM1 | SARM1 | 824 | 1.700 | 0.2351 | Yes |
| 145 | TNR | TNR | 837 | 1.676 | 0.2355 | Yes |
| 146 | SPTB | SPTB | 839 | 1.674 | 0.2365 | Yes |
| 147 | GDAP1L1 | GDAP1L1 | 849 | 1.667 | 0.2370 | Yes |
| 148 | AGT | AGT | 850 | 1.666 | 0.2380 | Yes |
| 149 | PPIL6 | PPIL6 | 851 | 1.666 | 0.2390 | Yes |
| 150 | NHLH1 | NHLH1 | 854 | 1.665 | 0.2400 | Yes |
| 151 | FAT3 | FAT3 | 858 | 1.663 | 0.2408 | Yes |
| 152 | IGSF11 | IGSF11 | 866 | 1.647 | 0.2415 | Yes |
| 153 | POU4F1 | POU4F1 | 874 | 1.640 | 0.2421 | Yes |
| 154 | DLGAP3 | DLGAP3 | 879 | 1.631 | 0.2429 | Yes |
| 155 | EPM2A | EPM2A | 882 | 1.629 | 0.2438 | Yes |
| 156 | PCDHB4 | PCDHB4 | 883 | 1.626 | 0.2448 | Yes |
| 157 | MORN5 | MORN5 | 885 | 1.622 | 0.2457 | Yes |
| 158 | PPFIA3 | PPFIA3 | 894 | 1.617 | 0.2463 | Yes |
| 159 | PRR36 | PRR36 | 896 | 1.613 | 0.2472 | Yes |
| 160 | ADAM11 | ADAM11 | 897 | 1.613 | 0.2482 | Yes |
| 161 | PAG1 | PAG1 | 898 | 1.613 | 0.2492 | Yes |
| 162 | NOVA2 | NOVA2 | 899 | 1.613 | 0.2502 | Yes |
| 163 | GAD1 | GAD1 | 905 | 1.603 | 0.2509 | Yes |
| 164 | VAX2 | VAX2 | 909 | 1.595 | 0.2517 | Yes |
| 165 | SLC26A8 | SLC26A8 | 914 | 1.584 | 0.2525 | Yes |
| 166 | PCDH1 | PCDH1 | 915 | 1.584 | 0.2534 | Yes |
| 167 | B3GAT2 | B3GAT2 | 917 | 1.579 | 0.2544 | Yes |
| 168 | RNASE1 | RNASE1 | 919 | 1.575 | 0.2553 | Yes |
| 169 | SYN1 | SYN1 | 920 | 1.574 | 0.2562 | Yes |
| 170 | OMG | OMG | 923 | 1.571 | 0.2571 | Yes |
| 171 | FCHO1 | FCHO1 | 928 | 1.566 | 0.2578 | Yes |
| 172 | TAGLN3 | TAGLN3 | 930 | 1.562 | 0.2587 | Yes |
| 173 | ZKSCAN4 | ZKSCAN4 | 943 | 1.552 | 0.2591 | Yes |
| 174 | SLC6A11 | SLC6A11 | 951 | 1.539 | 0.2596 | Yes |
| 175 | YPEL1 | YPEL1 | 953 | 1.532 | 0.2605 | Yes |
| 176 | SLC2A13 | SLC2A13 | 956 | 1.526 | 0.2614 | Yes |
| 177 | SLC29A4 | SLC29A4 | 957 | 1.524 | 0.2623 | Yes |
| 178 | TNFRSF19 | TNFRSF19 | 963 | 1.519 | 0.2630 | Yes |
| 179 | DUSP26 | DUSP26 | 969 | 1.509 | 0.2636 | Yes |
| 180 | MZB1 | MZB1 | 970 | 1.507 | 0.2645 | Yes |
| 181 | MANEAL | MANEAL | 972 | 1.505 | 0.2654 | Yes |
| 182 | NKAIN4 | NKAIN4 | 977 | 1.498 | 0.2661 | Yes |
| 183 | UNC13A | UNC13A | 978 | 1.496 | 0.2670 | Yes |
| 184 | TMEM145 | TMEM145 | 979 | 1.495 | 0.2679 | Yes |
| 185 | APBA2 | APBA2 | 984 | 1.493 | 0.2686 | Yes |
| 186 | PLXNB3 | PLXNB3 | 985 | 1.493 | 0.2696 | Yes |
| 187 | BCHE | BCHE | 988 | 1.489 | 0.2704 | Yes |
| 188 | IGFBPL1 | IGFBPL1 | 990 | 1.485 | 0.2712 | Yes |
| 189 | JPH3 | JPH3 | 991 | 1.483 | 0.2721 | Yes |
| 190 | CRB1 | CRB1 | 992 | 1.482 | 0.2730 | Yes |
| 191 | SAPCD2 | SAPCD2 | 1009 | 1.464 | 0.2731 | Yes |
| 192 | REM2 | REM2 | 1018 | 1.456 | 0.2736 | Yes |
| 193 | ACSS1 | ACSS1 | 1024 | 1.447 | 0.2742 | Yes |
| 194 | MFNG | MFNG | 1025 | 1.445 | 0.2751 | Yes |
| 195 | GRIK3 | GRIK3 | 1033 | 1.434 | 0.2756 | Yes |
| 196 | SPATA2L | SPATA2L | 1036 | 1.429 | 0.2764 | Yes |
| 197 | NOTCH4 | NOTCH4 | 1037 | 1.429 | 0.2772 | Yes |
| 198 | IGSF21 | IGSF21 | 1038 | 1.428 | 0.2781 | Yes |
| 199 | PPM1E | PPM1E | 1046 | 1.420 | 0.2786 | Yes |
| 200 | CMTM5 | CMTM5 | 1048 | 1.417 | 0.2794 | Yes |
| 201 | ADRA1A | ADRA1A | 1052 | 1.411 | 0.2801 | Yes |
| 202 | ASRGL1 | ASRGL1 | 1055 | 1.410 | 0.2809 | Yes |
| 203 | CADM2 | CADM2 | 1057 | 1.409 | 0.2817 | Yes |
| 204 | GPR162 | GPR162 | 1060 | 1.405 | 0.2825 | Yes |
| 205 | PHKG1 | PHKG1 | 1063 | 1.402 | 0.2832 | Yes |
| 206 | ELAVL3 | ELAVL3 | 1068 | 1.396 | 0.2839 | Yes |
| 207 | PCDHB11 | PCDHB11 | 1085 | 1.388 | 0.2839 | Yes |
| 208 | RNF144A | RNF144A | 1088 | 1.386 | 0.2846 | Yes |
| 209 | NCKAP5 | NCKAP5 | 1089 | 1.386 | 0.2855 | Yes |
| 210 | RHBDL3 | RHBDL3 | 1094 | 1.381 | 0.2861 | Yes |
| 211 | DPYSL5 | DPYSL5 | 1099 | 1.375 | 0.2868 | Yes |
| 212 | UNC5A | UNC5A | 1108 | 1.366 | 0.2872 | Yes |
| 213 | PLEKHH1 | PLEKHH1 | 1117 | 1.362 | 0.2876 | Yes |
| 214 | ATP6V1G2 | ATP6V1G2 | 1120 | 1.358 | 0.2883 | Yes |
| 215 | ADCYAP1R1 | ADCYAP1R1 | 1127 | 1.351 | 0.2889 | Yes |
| 216 | SOX6 | SOX6 | 1128 | 1.349 | 0.2897 | Yes |
| 217 | ASPRV1 | ASPRV1 | 1134 | 1.343 | 0.2902 | Yes |
| 218 | SHISA7 | SHISA7 | 1136 | 1.343 | 0.2910 | Yes |
| 219 | SLC27A5 | SLC27A5 | 1138 | 1.343 | 0.2918 | Yes |
| 220 | ANKRD44 | ANKRD44 | 1141 | 1.342 | 0.2925 | Yes |
| 221 | CHRNA4 | CHRNA4 | 1146 | 1.335 | 0.2931 | Yes |
| 222 | PRODH | PRODH | 1150 | 1.333 | 0.2938 | Yes |
| 223 | SNCAIP | SNCAIP | 1153 | 1.331 | 0.2945 | Yes |
| 224 | YPEL4 | YPEL4 | 1161 | 1.328 | 0.2949 | Yes |
| 225 | ST3GAL6 | ST3GAL6 | 1162 | 1.328 | 0.2957 | Yes |
| 226 | LRRN2 | LRRN2 | 1180 | 1.313 | 0.2957 | Yes |
| 227 | MMD2 | MMD2 | 1182 | 1.312 | 0.2964 | Yes |
| 228 | HCN3 | HCN3 | 1183 | 1.312 | 0.2972 | Yes |
| 229 | INHBB | INHBB | 1186 | 1.306 | 0.2979 | Yes |
| 230 | KCNIP2 | KCNIP2 | 1187 | 1.306 | 0.2987 | Yes |
| 231 | RIMKLB | RIMKLB | 1197 | 1.295 | 0.2990 | Yes |
| 232 | NXPH1 | NXPH1 | 1205 | 1.286 | 0.2995 | Yes |
| 233 | SNX22 | SNX22 | 1212 | 1.275 | 0.2999 | Yes |
| 234 | ABAT | ABAT | 1213 | 1.274 | 0.3007 | Yes |
| 235 | SYP | SYP | 1223 | 1.268 | 0.3010 | Yes |
| 236 | MYO16 | MYO16 | 1224 | 1.267 | 0.3018 | Yes |
| 237 | PCSK6 | PCSK6 | 1227 | 1.264 | 0.3025 | Yes |
| 238 | GPR19 | GPR19 | 1234 | 1.258 | 0.3029 | Yes |
| 239 | STMN3 | STMN3 | 1237 | 1.256 | 0.3036 | Yes |
| 240 | BTN1A1 | BTN1A1 | 1246 | 1.250 | 0.3040 | Yes |
| 241 | PTPRN2 | PTPRN2 | 1248 | 1.250 | 0.3047 | Yes |
| 242 | ARX | ARX | 1257 | 1.249 | 0.3050 | Yes |
| 243 | LRRC4B | LRRC4B | 1258 | 1.249 | 0.3058 | Yes |
| 244 | FAM69C | FAM69C | 1262 | 1.247 | 0.3064 | Yes |
| 245 | HEY1 | HEY1 | 1266 | 1.246 | 0.3070 | Yes |
| 246 | NCAM2 | NCAM2 | 1267 | 1.242 | 0.3077 | Yes |
| 247 | CD247 | CD247 | 1269 | 1.242 | 0.3085 | Yes |
| 248 | FAM89A | FAM89A | 1271 | 1.242 | 0.3092 | Yes |
| 249 | MYH7 | MYH7 | 1272 | 1.242 | 0.3099 | Yes |
| 250 | KBTBD11 | KBTBD11 | 1281 | 1.238 | 0.3103 | Yes |
| 251 | NRARP | NRARP | 1284 | 1.231 | 0.3109 | Yes |
| 252 | EDNRB | EDNRB | 1286 | 1.230 | 0.3116 | Yes |
| 253 | TTYH1 | TTYH1 | 1289 | 1.228 | 0.3123 | Yes |
| 254 | CTTNBP2 | CTTNBP2 | 1297 | 1.223 | 0.3126 | Yes |
| 255 | ARHGDIG | ARHGDIG | 1298 | 1.222 | 0.3134 | Yes |
| 256 | NTRK2 | NTRK2 | 1299 | 1.221 | 0.3141 | Yes |
| 257 | NYNRIN | NYNRIN | 1304 | 1.220 | 0.3147 | Yes |
| 258 | FAM161A | FAM161A | 1307 | 1.215 | 0.3153 | Yes |
| 259 | NCALD | NCALD | 1316 | 1.210 | 0.3156 | Yes |
| 260 | FAM131B | FAM131B | 1325 | 1.204 | 0.3160 | Yes |
| 261 | PPP1R14C | PPP1R14C | 1332 | 1.198 | 0.3164 | Yes |
| 262 | GPR45 | GPR45 | 1338 | 1.194 | 0.3169 | Yes |
| 263 | FHIT | FHIT | 1342 | 1.194 | 0.3174 | Yes |
| 264 | NIM1K | NIM1K | 1345 | 1.192 | 0.3181 | Yes |
| 265 | LRRTM2 | LRRTM2 | 1347 | 1.186 | 0.3187 | Yes |
| 266 | CTXN1 | CTXN1 | 1354 | 1.180 | 0.3192 | Yes |
| 267 | TRO | TRO | 1358 | 1.178 | 0.3197 | Yes |
| 268 | RNFT2 | RNFT2 | 1366 | 1.173 | 0.3201 | Yes |
| 269 | UNC79 | UNC79 | 1367 | 1.173 | 0.3208 | Yes |
| 270 | PHYHIPL | PHYHIPL | 1368 | 1.172 | 0.3215 | Yes |
| 271 | TMEM151B | TMEM151B | 1369 | 1.172 | 0.3222 | Yes |
| 272 | MN1 | MN1 | 1372 | 1.170 | 0.3228 | Yes |
| 273 | LRRN1 | LRRN1 | 1373 | 1.170 | 0.3235 | Yes |
| 274 | HEPACAM | HEPACAM | 1374 | 1.170 | 0.3243 | Yes |
| 275 | ELMO1 | ELMO1 | 1386 | 1.166 | 0.3244 | Yes |
| 276 | ENHO | ENHO | 1387 | 1.165 | 0.3251 | Yes |
| 277 | SEZ6L | SEZ6L | 1388 | 1.163 | 0.3258 | Yes |
| 278 | NANOS3 | NANOS3 | 1389 | 1.162 | 0.3265 | Yes |
| 279 | FAM57B | FAM57B | 1394 | 1.161 | 0.3270 | Yes |
| 280 | PTPRD | PTPRD | 1398 | 1.160 | 0.3276 | Yes |
| 281 | ACBD7 | ACBD7 | 1400 | 1.159 | 0.3283 | Yes |
| 282 | SEPT3 | SEPT3 | 1404 | 1.157 | 0.3288 | Yes |
| 283 | LSAMP | LSAMP | 1410 | 1.153 | 0.3293 | Yes |
| 284 | ZSWIM5 | ZSWIM5 | 1416 | 1.149 | 0.3297 | Yes |
| 285 | LFNG | LFNG | 1417 | 1.149 | 0.3304 | Yes |
| 286 | PCDH15 | PCDH15 | 1420 | 1.146 | 0.3310 | Yes |
| 287 | MYT1 | MYT1 | 1426 | 1.144 | 0.3314 | Yes |
| 288 | CTNNA2 | CTNNA2 | 1430 | 1.143 | 0.3320 | Yes |
| 289 | MAGI2 | MAGI2 | 1431 | 1.142 | 0.3327 | Yes |
| 290 | PDE4B | PDE4B | 1434 | 1.142 | 0.3333 | Yes |
| 291 | BCAN | BCAN | 1442 | 1.138 | 0.3336 | Yes |
| 292 | SLC1A6 | SLC1A6 | 1445 | 1.135 | 0.3342 | Yes |
| 293 | MYBPC1 | MYBPC1 | 1447 | 1.135 | 0.3348 | Yes |
| 294 | SH3GL2 | SH3GL2 | 1464 | 1.132 | 0.3347 | Yes |
| 295 | TRIM67 | TRIM67 | 1470 | 1.129 | 0.3352 | Yes |
| 296 | PHF21B | PHF21B | 1478 | 1.122 | 0.3355 | Yes |
| 297 | FBLL1 | FBLL1 | 1479 | 1.122 | 0.3362 | Yes |
| 298 | CPNE5 | CPNE5 | 1493 | 1.113 | 0.3362 | Yes |
| 299 | WNT4 | WNT4 | 1494 | 1.113 | 0.3369 | Yes |
| 300 | MKRN3 | MKRN3 | 1504 | 1.107 | 0.3371 | Yes |
| 301 | SRL | SRL | 1507 | 1.106 | 0.3377 | Yes |
| 302 | BEND5 | BEND5 | 1510 | 1.104 | 0.3382 | Yes |
| 303 | CACNB1 | CACNB1 | 1513 | 1.101 | 0.3388 | Yes |
| 304 | LIFR | LIFR | 1518 | 1.098 | 0.3393 | Yes |
| 305 | ASIC1 | ASIC1 | 1519 | 1.098 | 0.3399 | Yes |
| 306 | SLITRK2 | SLITRK2 | 1520 | 1.097 | 0.3406 | Yes |
| 307 | ADGRG1 | ADGRG1 | 1528 | 1.094 | 0.3409 | Yes |
| 308 | TTC25 | TTC25 | 1532 | 1.091 | 0.3414 | Yes |
| 309 | PRKCG | PRKCG | 1536 | 1.091 | 0.3419 | Yes |
| 310 | SAMD14 | SAMD14 | 1541 | 1.090 | 0.3424 | Yes |
| 311 | CDKN1C | CDKN1C | 1542 | 1.089 | 0.3431 | Yes |
| 312 | KCNA2 | KCNA2 | 1546 | 1.087 | 0.3436 | Yes |
| 313 | RGS9BP | RGS9BP | 1550 | 1.086 | 0.3441 | Yes |
| 314 | SLC22A17 | SLC22A17 | 1551 | 1.085 | 0.3447 | Yes |
| 315 | GRIA2 | GRIA2 | 1556 | 1.083 | 0.3452 | Yes |
| 316 | CBX2 | CBX2 | 1558 | 1.082 | 0.3458 | Yes |
| 317 | RASL10B | RASL10B | 1571 | 1.078 | 0.3459 | Yes |
| 318 | SLC8A2 | SLC8A2 | 1576 | 1.075 | 0.3463 | Yes |
| 319 | DCX | DCX | 1578 | 1.074 | 0.3469 | Yes |
| 320 | SPARCL1 | SPARCL1 | 1588 | 1.068 | 0.3471 | Yes |
| 321 | NACAD | NACAD | 1597 | 1.064 | 0.3473 | Yes |
| 322 | RAMP2 | RAMP2 | 1601 | 1.060 | 0.3478 | Yes |
| 323 | TMPRSS5 | TMPRSS5 | 1619 | 1.052 | 0.3476 | Yes |
| 324 | MPPED2 | MPPED2 | 1626 | 1.047 | 0.3480 | Yes |
| 325 | PCDH10 | PCDH10 | 1628 | 1.046 | 0.3485 | Yes |
| 326 | ADGRB3 | ADGRB3 | 1629 | 1.046 | 0.3492 | Yes |
| 327 | HAUS5 | HAUS5 | 1631 | 1.041 | 0.3498 | Yes |
| 328 | AGAP2 | AGAP2 | 1640 | 1.036 | 0.3500 | Yes |
| 329 | SALL2 | SALL2 | 1644 | 1.036 | 0.3505 | Yes |
| 330 | CHN2 | CHN2 | 1647 | 1.035 | 0.3510 | Yes |
| 331 | GABRG1 | GABRG1 | 1648 | 1.034 | 0.3516 | Yes |
| 332 | DMXL2 | DMXL2 | 1649 | 1.034 | 0.3522 | Yes |
| 333 | PCDHB6 | PCDHB6 | 1651 | 1.034 | 0.3528 | Yes |
| 334 | RNF165 | RNF165 | 1657 | 1.031 | 0.3532 | Yes |
| 335 | CCDC106 | CCDC106 | 1662 | 1.027 | 0.3536 | Yes |
| 336 | GPR37L1 | GPR37L1 | 1665 | 1.027 | 0.3541 | Yes |
| 337 | MYCN | MYCN | 1667 | 1.026 | 0.3547 | Yes |
| 338 | FGF11 | FGF11 | 1669 | 1.025 | 0.3553 | Yes |
| 339 | ADGRB2 | ADGRB2 | 1671 | 1.023 | 0.3559 | Yes |
| 340 | PAQR4 | PAQR4 | 1672 | 1.022 | 0.3565 | Yes |
| 341 | MPP2 | MPP2 | 1673 | 1.022 | 0.3571 | Yes |
| 342 | SBK1 | SBK1 | 1682 | 1.016 | 0.3573 | Yes |
| 343 | TPPP | TPPP | 1684 | 1.015 | 0.3579 | Yes |
| 344 | CADM4 | CADM4 | 1692 | 1.012 | 0.3582 | Yes |
| 345 | MRO | MRO | 1695 | 1.010 | 0.3587 | Yes |
| 346 | CCDC150 | CCDC150 | 1700 | 1.008 | 0.3591 | Yes |
| 347 | PTPRZ1 | PTPRZ1 | 1701 | 1.008 | 0.3597 | Yes |
| 348 | SEMA6C | SEMA6C | 1705 | 1.004 | 0.3602 | Yes |
| 349 | RFTN2 | RFTN2 | 1706 | 1.004 | 0.3608 | Yes |
| 350 | DLL3 | DLL3 | 1711 | 1.002 | 0.3612 | Yes |
| 351 | MAP2 | MAP2 | 1712 | 1.001 | 0.3618 | Yes |
| 352 | HAP1 | HAP1 | 1715 | 0.998 | 0.3623 | Yes |
| 353 | TRIM71 | TRIM71 | 1719 | 0.997 | 0.3627 | Yes |
| 354 | MYCL | MYCL | 1720 | 0.997 | 0.3634 | Yes |
| 355 | HES5 | HES5 | 1722 | 0.996 | 0.3639 | Yes |
| 356 | MTERF2 | MTERF2 | 1723 | 0.995 | 0.3645 | Yes |
| 357 | N4BP2L1 | N4BP2L1 | 1728 | 0.992 | 0.3649 | Yes |
| 358 | TMEM198 | TMEM198 | 1733 | 0.991 | 0.3653 | Yes |
| 359 | B3GAT1 | B3GAT1 | 1738 | 0.988 | 0.3657 | Yes |
| 360 | SCRG1 | SCRG1 | 1740 | 0.986 | 0.3663 | Yes |
| 361 | CAMKV | CAMKV | 1745 | 0.983 | 0.3667 | Yes |
| 362 | GABRB3 | GABRB3 | 1749 | 0.981 | 0.3671 | Yes |
| 363 | GPM6A | GPM6A | 1751 | 0.979 | 0.3677 | Yes |
| 364 | TMIE | TMIE | 1754 | 0.978 | 0.3681 | Yes |
| 365 | DDX4 | DDX4 | 1759 | 0.975 | 0.3685 | Yes |
| 366 | CAMK2A | CAMK2A | 1763 | 0.974 | 0.3690 | Yes |
| 367 | TBX6 | TBX6 | 1767 | 0.972 | 0.3694 | Yes |
| 368 | SOX8 | SOX8 | 1771 | 0.971 | 0.3699 | Yes |
| 369 | SMOC1 | SMOC1 | 1772 | 0.971 | 0.3704 | Yes |
| 370 | RASSF2 | RASSF2 | 1773 | 0.970 | 0.3710 | Yes |
| 371 | ILDR2 | ILDR2 | 1775 | 0.970 | 0.3716 | Yes |
| 372 | LRP3 | LRP3 | 1782 | 0.964 | 0.3719 | Yes |
| 373 | ZFP14 | ZFP14 | 1783 | 0.964 | 0.3724 | Yes |
| 374 | SHANK1 | SHANK1 | 1790 | 0.963 | 0.3727 | Yes |
| 375 | DFFB | DFFB | 1791 | 0.963 | 0.3733 | Yes |
| 376 | NOXO1 | NOXO1 | 1792 | 0.962 | 0.3739 | Yes |
| 377 | TUBB2B | TUBB2B | 1795 | 0.961 | 0.3744 | Yes |
| 378 | RTN1 | RTN1 | 1805 | 0.958 | 0.3745 | Yes |
| 379 | TBC1D16 | TBC1D16 | 1807 | 0.953 | 0.3750 | Yes |
| 380 | AQP4 | AQP4 | 1809 | 0.953 | 0.3756 | Yes |
| 381 | CNTFR | CNTFR | 1813 | 0.952 | 0.3760 | Yes |
| 382 | KCNQ3 | KCNQ3 | 1815 | 0.950 | 0.3765 | Yes |
| 383 | HAGHL | HAGHL | 1820 | 0.948 | 0.3769 | Yes |
| 384 | GRIA4 | GRIA4 | 1825 | 0.945 | 0.3773 | Yes |
| 385 | ERBB4 | ERBB4 | 1826 | 0.945 | 0.3779 | Yes |
| 386 | ABCB6 | ABCB6 | 1833 | 0.943 | 0.3781 | Yes |
| 387 | SEMA4G | SEMA4G | 1836 | 0.942 | 0.3786 | Yes |
| 388 | PIK3C2B | PIK3C2B | 1839 | 0.940 | 0.3791 | Yes |
| 389 | APBA1 | APBA1 | 1841 | 0.939 | 0.3796 | Yes |
| 390 | PTCH1 | PTCH1 | 1842 | 0.938 | 0.3802 | Yes |
| 391 | RALGPS1 | RALGPS1 | 1848 | 0.933 | 0.3805 | Yes |
| 392 | PCDHB12 | PCDHB12 | 1849 | 0.932 | 0.3810 | Yes |
| 393 | NDRG2 | NDRG2 | 1850 | 0.932 | 0.3816 | Yes |
| 394 | MYO10 | MYO10 | 1853 | 0.931 | 0.3821 | Yes |
| 395 | CHRM1 | CHRM1 | 1858 | 0.929 | 0.3824 | Yes |
| 396 | LYSMD1 | LYSMD1 | 1859 | 0.927 | 0.3830 | Yes |
| 397 | PPFIBP2 | PPFIBP2 | 1864 | 0.924 | 0.3834 | Yes |
| 398 | LGR5 | LGR5 | 1868 | 0.922 | 0.3838 | Yes |
| 399 | ZBTB3 | ZBTB3 | 1874 | 0.920 | 0.3841 | Yes |
| 400 | GPR155 | GPR155 | 1878 | 0.919 | 0.3845 | Yes |
| 401 | ACSBG1 | ACSBG1 | 1879 | 0.919 | 0.3850 | Yes |
| 402 | HRH3 | HRH3 | 1886 | 0.917 | 0.3853 | Yes |
| 403 | MAPT | MAPT | 1887 | 0.917 | 0.3859 | Yes |
| 404 | SHANK2 | SHANK2 | 1898 | 0.912 | 0.3859 | Yes |
| 405 | FAM229B | FAM229B | 1903 | 0.909 | 0.3863 | Yes |
| 406 | NKD1 | NKD1 | 1911 | 0.906 | 0.3865 | Yes |
| 407 | KCNN2 | KCNN2 | 1914 | 0.903 | 0.3869 | Yes |
| 408 | MLC1 | MLC1 | 1922 | 0.899 | 0.3871 | Yes |
| 409 | TSPAN12 | TSPAN12 | 1927 | 0.897 | 0.3874 | Yes |
| 410 | BTG2 | BTG2 | 1929 | 0.896 | 0.3879 | Yes |
| 411 | COL16A1 | COL16A1 | 1932 | 0.893 | 0.3884 | Yes |
| 412 | MAD2L2 | MAD2L2 | 1933 | 0.891 | 0.3889 | Yes |
| 413 | KBTBD7 | KBTBD7 | 1946 | 0.886 | 0.3889 | Yes |
| 414 | CPEB2 | CPEB2 | 1949 | 0.885 | 0.3893 | Yes |
| 415 | NREP | NREP | 1955 | 0.881 | 0.3896 | Yes |
| 416 | PSD | PSD | 1960 | 0.878 | 0.3899 | Yes |
| 417 | KIFC2 | KIFC2 | 1972 | 0.873 | 0.3899 | Yes |
| 418 | ERICH2 | ERICH2 | 1973 | 0.872 | 0.3904 | Yes |
| 419 | TXNDC16 | TXNDC16 | 1976 | 0.872 | 0.3908 | Yes |
| 420 | TCEA2 | TCEA2 | 1977 | 0.871 | 0.3914 | Yes |
| 421 | SCAMP5 | SCAMP5 | 1980 | 0.869 | 0.3918 | Yes |
| 422 | OLIG2 | OLIG2 | 1982 | 0.868 | 0.3923 | Yes |
| 423 | PPP1R3E | PPP1R3E | 1986 | 0.866 | 0.3926 | Yes |
| 424 | KLF15 | KLF15 | 1988 | 0.865 | 0.3931 | Yes |
| 425 | ALDH6A1 | ALDH6A1 | 1993 | 0.864 | 0.3934 | Yes |
| 426 | KCND2 | KCND2 | 1995 | 0.863 | 0.3939 | Yes |
| 427 | MEIS1 | MEIS1 | 2001 | 0.860 | 0.3942 | Yes |
| 428 | GAB1 | GAB1 | 2006 | 0.859 | 0.3945 | Yes |
| 429 | SASH1 | SASH1 | 2015 | 0.856 | 0.3946 | Yes |
| 430 | KCND3 | KCND3 | 2021 | 0.854 | 0.3949 | Yes |
| 431 | DBNDD2 | DBNDD2 | 2022 | 0.853 | 0.3954 | Yes |
| 432 | EEPD1 | EEPD1 | 2026 | 0.851 | 0.3958 | Yes |
| 433 | BMP7 | BMP7 | 2038 | 0.845 | 0.3957 | Yes |
| 434 | PLLP | PLLP | 2039 | 0.845 | 0.3963 | Yes |
| 435 | HFM1 | HFM1 | 2041 | 0.845 | 0.3967 | Yes |
| 436 | DPYSL4 | DPYSL4 | 2045 | 0.842 | 0.3971 | Yes |
| 437 | CACNA1H | CACNA1H | 2046 | 0.842 | 0.3976 | Yes |
| 438 | EGFR | EGFR | 2052 | 0.839 | 0.3978 | Yes |
| 439 | DCLK2 | DCLK2 | 2054 | 0.838 | 0.3983 | Yes |
| 440 | BACH2 | BACH2 | 2057 | 0.836 | 0.3987 | Yes |
| 441 | FRMPD1 | FRMPD1 | 2060 | 0.834 | 0.3991 | Yes |
| 442 | PGAP1 | PGAP1 | 2063 | 0.833 | 0.3995 | Yes |
| 443 | DDHD1 | DDHD1 | 2065 | 0.833 | 0.4000 | Yes |
| 444 | ATP1A3 | ATP1A3 | 2066 | 0.833 | 0.4005 | Yes |
| 445 | PTPRO | PTPRO | 2068 | 0.832 | 0.4009 | Yes |
| 446 | CBS | CBS | 2071 | 0.831 | 0.4014 | Yes |
| 447 | CCDC28B | CCDC28B | 2083 | 0.827 | 0.4013 | Yes |
| 448 | PNCK | PNCK | 2084 | 0.827 | 0.4018 | Yes |
| 449 | CIRBP | CIRBP | 2089 | 0.824 | 0.4021 | Yes |
| 450 | EFNA2 | EFNA2 | 2091 | 0.823 | 0.4026 | Yes |
| 451 | SV2A | SV2A | 2099 | 0.820 | 0.4027 | Yes |
| 452 | TMEM136 | TMEM136 | 2102 | 0.819 | 0.4031 | Yes |
| 453 | NHSL1 | NHSL1 | 2103 | 0.818 | 0.4036 | Yes |
| 454 | ZSCAN22 | ZSCAN22 | 2104 | 0.818 | 0.4041 | Yes |
| 455 | CECR2 | CECR2 | 2106 | 0.818 | 0.4045 | Yes |
| 456 | PTPRT | PTPRT | 2108 | 0.817 | 0.4050 | Yes |
| 457 | COLGALT2 | COLGALT2 | 2111 | 0.816 | 0.4054 | Yes |
| 458 | DISC1 | DISC1 | 2114 | 0.815 | 0.4058 | Yes |
| 459 | NCAN | NCAN | 2117 | 0.813 | 0.4062 | Yes |
| 460 | EFCAB7 | EFCAB7 | 2118 | 0.813 | 0.4067 | Yes |
| 461 | NUDT11 | NUDT11 | 2119 | 0.813 | 0.4072 | Yes |
| 462 | ARHGEF9 | ARHGEF9 | 2122 | 0.810 | 0.4076 | Yes |
| 463 | ATP1B2 | ATP1B2 | 2123 | 0.810 | 0.4080 | Yes |
| 464 | KY | KY | 2124 | 0.810 | 0.4085 | Yes |
| 465 | CTNND2 | CTNND2 | 2127 | 0.809 | 0.4089 | Yes |
| 466 | SLC38A3 | SLC38A3 | 2128 | 0.809 | 0.4094 | Yes |
| 467 | PXMP2 | PXMP2 | 2132 | 0.808 | 0.4098 | Yes |
| 468 | CLEC11A | CLEC11A | 2133 | 0.807 | 0.4103 | Yes |
| 469 | STX1B | STX1B | 2137 | 0.805 | 0.4106 | Yes |
| 470 | TIMP4 | TIMP4 | 2138 | 0.805 | 0.4111 | Yes |
| 471 | DLX1 | DLX1 | 2139 | 0.804 | 0.4116 | Yes |
| 472 | NSUN6 | NSUN6 | 2142 | 0.802 | 0.4120 | Yes |
| 473 | MEIS3 | MEIS3 | 2150 | 0.801 | 0.4121 | Yes |
| 474 | AMER2 | AMER2 | 2157 | 0.798 | 0.4123 | Yes |
| 475 | OLIG1 | OLIG1 | 2160 | 0.796 | 0.4127 | Yes |
| 476 | NOTCH3 | NOTCH3 | 2163 | 0.795 | 0.4130 | Yes |
| 477 | FAM71E1 | FAM71E1 | 2164 | 0.794 | 0.4135 | Yes |
| 478 | SOX21 | SOX21 | 2169 | 0.793 | 0.4138 | Yes |
| 479 | CSPG5 | CSPG5 | 2171 | 0.792 | 0.4142 | Yes |
| 480 | LGALSL | LGALSL | 2173 | 0.792 | 0.4147 | Yes |
| 481 | DLL1 | DLL1 | 2175 | 0.790 | 0.4151 | Yes |
| 482 | MARCKSL1 | MARCKSL1 | 2177 | 0.790 | 0.4155 | Yes |
| 483 | CHADL | CHADL | 2184 | 0.789 | 0.4157 | Yes |
| 484 | TCEANC | TCEANC | 2189 | 0.786 | 0.4160 | Yes |
| 485 | KISS1R | KISS1R | 2191 | 0.785 | 0.4164 | Yes |
| 486 | SLC1A3 | SLC1A3 | 2195 | 0.783 | 0.4167 | Yes |
| 487 | NDRG4 | NDRG4 | 2198 | 0.782 | 0.4171 | Yes |
| 488 | PPP1R3C | PPP1R3C | 2203 | 0.778 | 0.4174 | Yes |
| 489 | TRAF4 | TRAF4 | 2204 | 0.776 | 0.4179 | Yes |
| 490 | FBXO21 | FBXO21 | 2205 | 0.775 | 0.4183 | Yes |
| 491 | LHFPL3 | LHFPL3 | 2209 | 0.773 | 0.4186 | Yes |
| 492 | ZSCAN21 | ZSCAN21 | 2214 | 0.771 | 0.4189 | Yes |
| 493 | REV3L | REV3L | 2219 | 0.769 | 0.4192 | Yes |
| 494 | IGLON5 | IGLON5 | 2225 | 0.767 | 0.4194 | Yes |
| 495 | ARMCX5 | ARMCX5 | 2226 | 0.766 | 0.4199 | Yes |
| 496 | KIZ | KIZ | 2231 | 0.765 | 0.4201 | Yes |
| 497 | ASB4 | ASB4 | 2232 | 0.765 | 0.4206 | Yes |
| 498 | MTURN | MTURN | 2236 | 0.763 | 0.4209 | Yes |
| 499 | SNN | SNN | 2242 | 0.761 | 0.4211 | Yes |
| 500 | KIF1A | KIF1A | 2255 | 0.757 | 0.4210 | Yes |
| 501 | DPF1 | DPF1 | 2257 | 0.756 | 0.4214 | Yes |
| 502 | ZFP37 | ZFP37 | 2263 | 0.753 | 0.4216 | Yes |
| 503 | EFNB3 | EFNB3 | 2268 | 0.752 | 0.4218 | Yes |
| 504 | MAP4K1 | MAP4K1 | 2270 | 0.751 | 0.4222 | Yes |
| 505 | PLP1 | PLP1 | 2275 | 0.750 | 0.4225 | Yes |
| 506 | IQCC | IQCC | 2287 | 0.747 | 0.4224 | Yes |
| 507 | RFX3 | RFX3 | 2288 | 0.746 | 0.4228 | Yes |
| 508 | SOX5 | SOX5 | 2291 | 0.745 | 0.4232 | Yes |
| 509 | KIF21A | KIF21A | 2295 | 0.744 | 0.4235 | Yes |
| 510 | ZBTB39 | ZBTB39 | 2305 | 0.740 | 0.4235 | Yes |
| 511 | PCMTD2 | PCMTD2 | 2314 | 0.738 | 0.4235 | Yes |
| 512 | SRGAP3 | SRGAP3 | 2320 | 0.737 | 0.4237 | Yes |
| 513 | FAM228B | FAM228B | 2323 | 0.736 | 0.4241 | Yes |
| 514 | ASIC3 | ASIC3 | 2325 | 0.736 | 0.4245 | Yes |
| 515 | GBA2 | GBA2 | 2329 | 0.735 | 0.4248 | Yes |
| 516 | HSF2BP | HSF2BP | 2330 | 0.735 | 0.4252 | Yes |
| 517 | ITGAL | ITGAL | 2332 | 0.735 | 0.4256 | Yes |
| 518 | TMEM59L | TMEM59L | 2333 | 0.735 | 0.4261 | Yes |
| 519 | KLHL11 | KLHL11 | 2352 | 0.734 | 0.4256 | Yes |
| 520 | CDK5R1 | CDK5R1 | 2357 | 0.734 | 0.4258 | Yes |
| 521 | CACNG8 | CACNG8 | 2359 | 0.733 | 0.4262 | Yes |
| 522 | SLC25A53 | SLC25A53 | 2390 | 0.730 | 0.4252 | Yes |
| 523 | ALKBH4 | ALKBH4 | 2399 | 0.730 | 0.4252 | Yes |
| 524 | NFIA | NFIA | 2401 | 0.730 | 0.4256 | Yes |
| 525 | SRSF12 | SRSF12 | 2412 | 0.728 | 0.4255 | Yes |
| 526 | ATP1A2 | ATP1A2 | 2413 | 0.728 | 0.4260 | Yes |
| 527 | CLASP2 | CLASP2 | 2418 | 0.727 | 0.4262 | Yes |
| 528 | FA2H | FA2H | 2419 | 0.727 | 0.4267 | Yes |
| 529 | KCTD14 | KCTD14 | 2422 | 0.727 | 0.4270 | Yes |
| 530 | ACVR2B | ACVR2B | 2451 | 0.724 | 0.4260 | Yes |
| 531 | SYNGR3 | SYNGR3 | 2457 | 0.723 | 0.4262 | Yes |
| 532 | ARAP2 | ARAP2 | 2460 | 0.722 | 0.4266 | Yes |
| 533 | RPH3AL | RPH3AL | 2471 | 0.719 | 0.4265 | Yes |
| 534 | GRM3 | GRM3 | 2477 | 0.715 | 0.4267 | Yes |
| 535 | FZD3 | FZD3 | 2478 | 0.715 | 0.4271 | Yes |
| 536 | BTBD17 | BTBD17 | 2483 | 0.713 | 0.4273 | Yes |
| 537 | ADCY5 | ADCY5 | 2484 | 0.712 | 0.4278 | Yes |
| 538 | SLC25A23 | SLC25A23 | 2488 | 0.710 | 0.4280 | Yes |
| 539 | NAT8L | NAT8L | 2495 | 0.706 | 0.4282 | Yes |
| 540 | REEP1 | REEP1 | 2501 | 0.706 | 0.4284 | Yes |
| 541 | PURG | PURG | 2533 | 0.704 | 0.4272 | Yes |
| 542 | PCDHGC3 | PCDHGC3 | 2540 | 0.703 | 0.4273 | Yes |
| 543 | PLXNB1 | PLXNB1 | 2573 | 0.699 | 0.4261 | Yes |
| 544 | SH3YL1 | SH3YL1 | 2575 | 0.699 | 0.4265 | Yes |
| 545 | PSPC1 | PSPC1 | 2576 | 0.699 | 0.4269 | Yes |
| 546 | ZBTB34 | ZBTB34 | 2577 | 0.698 | 0.4274 | Yes |
| 547 | SKOR1 | SKOR1 | 2578 | 0.698 | 0.4278 | Yes |
| 548 | KLHL7 | KLHL7 | 2579 | 0.697 | 0.4282 | Yes |
| 549 | ZSCAN29 | ZSCAN29 | 2587 | 0.696 | 0.4283 | Yes |
| 550 | BCL7A | BCL7A | 2589 | 0.695 | 0.4287 | Yes |
| 551 | NOVA1 | NOVA1 | 2593 | 0.694 | 0.4289 | Yes |
| 552 | CITED1 | CITED1 | 2597 | 0.692 | 0.4292 | Yes |
| 553 | ZKSCAN2 | ZKSCAN2 | 2614 | 0.687 | 0.4288 | Yes |
| 554 | FARP2 | FARP2 | 2617 | 0.686 | 0.4291 | Yes |
| 555 | MAGEL2 | MAGEL2 | 2620 | 0.686 | 0.4294 | Yes |
| 556 | APOE | APOE | 2626 | 0.685 | 0.4296 | Yes |
| 557 | CHST11 | CHST11 | 2627 | 0.685 | 0.4300 | Yes |
| 558 | TBKBP1 | TBKBP1 | 2630 | 0.682 | 0.4303 | Yes |
| 559 | DOCK3 | DOCK3 | 2631 | 0.682 | 0.4307 | Yes |
| 560 | CCDC173 | CCDC173 | 2633 | 0.681 | 0.4311 | Yes |
| 561 | SYNJ1 | SYNJ1 | 2635 | 0.681 | 0.4315 | Yes |
| 562 | RUFY3 | RUFY3 | 2645 | 0.679 | 0.4314 | Yes |
| 563 | TMEM74B | TMEM74B | 2646 | 0.679 | 0.4318 | Yes |
| 564 | AXIN2 | AXIN2 | 2649 | 0.678 | 0.4322 | Yes |
| 565 | CSGALNACT1 | CSGALNACT1 | 2651 | 0.677 | 0.4325 | Yes |
| 566 | DHX35 | DHX35 | 2654 | 0.676 | 0.4328 | Yes |
| 567 | ABCD2 | ABCD2 | 2655 | 0.676 | 0.4332 | Yes |
| 568 | PDZD4 | PDZD4 | 2656 | 0.676 | 0.4337 | Yes |
| 569 | MADD | MADD | 2661 | 0.675 | 0.4339 | Yes |
| 570 | RBMX | RBMX | 2662 | 0.675 | 0.4343 | Yes |
| 571 | FBXL16 | FBXL16 | 2664 | 0.674 | 0.4346 | Yes |
| 572 | PCCB | PCCB | 2666 | 0.673 | 0.4350 | Yes |
| 573 | MAP6D1 | MAP6D1 | 2679 | 0.671 | 0.4348 | Yes |
| 574 | DDN | DDN | 2685 | 0.671 | 0.4349 | Yes |
| 575 | ZKSCAN5 | ZKSCAN5 | 2712 | 0.671 | 0.4340 | Yes |
| 576 | SLC4A4 | SLC4A4 | 2714 | 0.670 | 0.4344 | Yes |
| 577 | SOX11 | SOX11 | 2717 | 0.668 | 0.4347 | Yes |
| 578 | RFX2 | RFX2 | 2718 | 0.667 | 0.4351 | Yes |
| 579 | TBC1D14 | TBC1D14 | 2719 | 0.667 | 0.4355 | Yes |
| 580 | GLT1D1 | GLT1D1 | 2735 | 0.663 | 0.4352 | Yes |
| 581 | FAM69B | FAM69B | 2740 | 0.661 | 0.4354 | Yes |
| 582 | FOXJ1 | FOXJ1 | 2742 | 0.661 | 0.4357 | Yes |
| 583 | PTN | PTN | 2750 | 0.659 | 0.4358 | Yes |
| 584 | CNPY1 | CNPY1 | 2760 | 0.656 | 0.4357 | Yes |
| 585 | PFN4 | PFN4 | 2762 | 0.656 | 0.4361 | Yes |
| 586 | SH2D4B | SH2D4B | 2773 | 0.654 | 0.4359 | Yes |
| 587 | CRIP3 | CRIP3 | 2780 | 0.654 | 0.4360 | Yes |
| 588 | AGER | AGER | 2784 | 0.653 | 0.4363 | Yes |
| 589 | SOBP | SOBP | 2786 | 0.653 | 0.4366 | Yes |
| 590 | FAM196A | FAM196A | 2815 | 0.650 | 0.4356 | Yes |
| 591 | KCNC3 | KCNC3 | 2823 | 0.649 | 0.4357 | Yes |
| 592 | MACROD2 | MACROD2 | 2826 | 0.649 | 0.4359 | Yes |
| 593 | FEZ1 | FEZ1 | 2830 | 0.647 | 0.4362 | Yes |
| 594 | GPT | GPT | 2831 | 0.646 | 0.4366 | Yes |
| 595 | WDR27 | WDR27 | 2844 | 0.643 | 0.4364 | Yes |
| 596 | CCER2 | CCER2 | 2853 | 0.643 | 0.4364 | Yes |
| 597 | CASQ1 | CASQ1 | 2862 | 0.643 | 0.4363 | Yes |
| 598 | SLC32A1 | SLC32A1 | 2867 | 0.643 | 0.4365 | Yes |
| 599 | ADGRG3 | ADGRG3 | 2872 | 0.643 | 0.4367 | Yes |
| 600 | GARNL3 | GARNL3 | 2885 | 0.643 | 0.4365 | Yes |
| 601 | SLC12A6 | SLC12A6 | 2887 | 0.643 | 0.4368 | Yes |
| 602 | POU6F1 | POU6F1 | 2903 | 0.640 | 0.4365 | Yes |
| 603 | GLUL | GLUL | 2904 | 0.639 | 0.4369 | Yes |
| 604 | MGAT4A | MGAT4A | 2905 | 0.639 | 0.4373 | Yes |
| 605 | ANKRD13B | ANKRD13B | 2906 | 0.639 | 0.4376 | Yes |
| 606 | SYN2 | SYN2 | 2910 | 0.638 | 0.4379 | Yes |
| 607 | ABLIM2 | ABLIM2 | 2913 | 0.638 | 0.4382 | Yes |
| 608 | FBXO24 | FBXO24 | 2915 | 0.638 | 0.4385 | Yes |
| 609 | ASCL1 | ASCL1 | 2920 | 0.636 | 0.4387 | Yes |
| 610 | NUPL2 | NUPL2 | 2923 | 0.635 | 0.4390 | Yes |
| 611 | NXPH3 | NXPH3 | 2931 | 0.634 | 0.4390 | Yes |
| 612 | SP8 | SP8 | 2933 | 0.634 | 0.4393 | Yes |
| 613 | TMPRSS9 | TMPRSS9 | 2936 | 0.634 | 0.4396 | Yes |
| 614 | RAP2A | RAP2A | 2962 | 0.633 | 0.4387 | Yes |
| 615 | LRRC57 | LRRC57 | 2971 | 0.633 | 0.4387 | Yes |
| 616 | RND2 | RND2 | 2974 | 0.632 | 0.4390 | Yes |
| 617 | STXBP5L | STXBP5L | 2985 | 0.632 | 0.4389 | Yes |
| 618 | EFHB | EFHB | 2986 | 0.632 | 0.4393 | Yes |
| 619 | ABCG1 | ABCG1 | 2987 | 0.632 | 0.4397 | Yes |
| 620 | RPH3A | RPH3A | 2993 | 0.632 | 0.4398 | Yes |
| 621 | ASIC4 | ASIC4 | 2999 | 0.632 | 0.4399 | Yes |
| 622 | CFAP61 | CFAP61 | 3004 | 0.632 | 0.4401 | Yes |
| 623 | GJB1 | GJB1 | 3009 | 0.632 | 0.4403 | Yes |
| 624 | MYO18B | MYO18B | 3021 | 0.632 | 0.4401 | Yes |
| 625 | NAPSA | NAPSA | 3081 | 0.631 | 0.4375 | Yes |
| 626 | SYT5 | SYT5 | 3082 | 0.631 | 0.4379 | Yes |
| 627 | ICA1L | ICA1L | 3086 | 0.631 | 0.4381 | Yes |
| 628 | BARHL1 | BARHL1 | 3087 | 0.631 | 0.4385 | Yes |
| 629 | USHBP1 | USHBP1 | 3100 | 0.631 | 0.4383 | Yes |
| 630 | TRPM8 | TRPM8 | 3102 | 0.631 | 0.4386 | Yes |
| 631 | NEUROD2 | NEUROD2 | 3106 | 0.631 | 0.4389 | Yes |
| 632 | IQUB | IQUB | 3113 | 0.631 | 0.4389 | Yes |
| 633 | GRID2 | GRID2 | 3130 | 0.631 | 0.4385 | Yes |
| 634 | CFAP126 | CFAP126 | 3199 | 0.631 | 0.4355 | Yes |
| 635 | PER3 | PER3 | 3206 | 0.629 | 0.4355 | Yes |
| 636 | SLC15A2 | SLC15A2 | 3207 | 0.628 | 0.4359 | Yes |
| 637 | JAM2 | JAM2 | 3211 | 0.627 | 0.4361 | Yes |
| 638 | CENPT | CENPT | 3214 | 0.624 | 0.4364 | Yes |
| 639 | CNBD2 | CNBD2 | 3227 | 0.621 | 0.4362 | Yes |
| 640 | RTKN | RTKN | 3228 | 0.621 | 0.4366 | Yes |
| 641 | LHFPL4 | LHFPL4 | 3232 | 0.620 | 0.4368 | Yes |
| 642 | B4GALNT4 | B4GALNT4 | 3234 | 0.619 | 0.4371 | Yes |
| 643 | MED12L | MED12L | 3235 | 0.619 | 0.4375 | Yes |
| 644 | GNG7 | GNG7 | 3240 | 0.618 | 0.4377 | Yes |
| 645 | FUT9 | FUT9 | 3243 | 0.617 | 0.4379 | Yes |
| 646 | ATP8A1 | ATP8A1 | 3244 | 0.617 | 0.4383 | Yes |
| 647 | COL28A1 | COL28A1 | 3245 | 0.617 | 0.4387 | Yes |
| 648 | PCDHGC4 | PCDHGC4 | 3248 | 0.616 | 0.4390 | Yes |
| 649 | LPAR4 | LPAR4 | 3249 | 0.615 | 0.4393 | Yes |
| 650 | EVL | EVL | 3258 | 0.613 | 0.4393 | Yes |
| 651 | SYNPO2L | SYNPO2L | 3260 | 0.612 | 0.4396 | Yes |
| 652 | DKKL1 | DKKL1 | 3261 | 0.612 | 0.4400 | Yes |
| 653 | FGFBP3 | FGFBP3 | 3281 | 0.608 | 0.4394 | Yes |
| 654 | LRRC73 | LRRC73 | 3288 | 0.605 | 0.4395 | Yes |
| 655 | WDPCP | WDPCP | 3294 | 0.604 | 0.4396 | Yes |
| 656 | ABCC5 | ABCC5 | 3298 | 0.604 | 0.4398 | Yes |
| 657 | OSGEPL1 | OSGEPL1 | 3301 | 0.603 | 0.4401 | Yes |
| 658 | MDFI | MDFI | 3302 | 0.603 | 0.4404 | Yes |
| 659 | GRID2IP | GRID2IP | 3314 | 0.602 | 0.4403 | Yes |
| 660 | SPATA25 | SPATA25 | 3318 | 0.602 | 0.4405 | Yes |
| 661 | RPS6KL1 | RPS6KL1 | 3319 | 0.602 | 0.4408 | Yes |
| 662 | PAH | PAH | 3323 | 0.602 | 0.4411 | Yes |
| 663 | SLC26A4 | SLC26A4 | 3325 | 0.602 | 0.4414 | Yes |
| 664 | HAPLN1 | HAPLN1 | 3331 | 0.602 | 0.4415 | Yes |
| 665 | EBF2 | EBF2 | 3334 | 0.602 | 0.4417 | Yes |
| 666 | CPNE6 | CPNE6 | 3337 | 0.602 | 0.4420 | Yes |
| 667 | RASAL1 | RASAL1 | 3357 | 0.602 | 0.4414 | No |
| 668 | GNAT1 | GNAT1 | 3370 | 0.602 | 0.4412 | No |
| 669 | PLK5 | PLK5 | 3375 | 0.602 | 0.4413 | No |
| 670 | KCNE2 | KCNE2 | 3389 | 0.602 | 0.4410 | No |
| 671 | CRYGA | CRYGA | 3401 | 0.602 | 0.4409 | No |
| 672 | LRRC3B | LRRC3B | 3404 | 0.602 | 0.4411 | No |
| 673 | C2CD4C | C2CD4C | 3408 | 0.602 | 0.4413 | No |
| 674 | PTGS1 | PTGS1 | 3411 | 0.602 | 0.4416 | No |
| 675 | FAM78A | FAM78A | 3424 | 0.602 | 0.4414 | No |
| 676 | PKDREJ | PKDREJ | 3428 | 0.602 | 0.4416 | No |
| 677 | FAM229A | FAM229A | 3497 | 0.602 | 0.4385 | No |
| 678 | U2AF1L4 | U2AF1L4 | 3498 | 0.602 | 0.4389 | No |
| 679 | DCST2 | DCST2 | 3724 | 0.602 | 0.4279 | No |
| 680 | MARVELD3 | MARVELD3 | 3729 | 0.602 | 0.4280 | No |
| 681 | CLHC1 | CLHC1 | 3741 | 0.602 | 0.4278 | No |
| 682 | RGS6 | RGS6 | 3745 | 0.602 | 0.4280 | No |
| 683 | F12 | F12 | 3749 | 0.602 | 0.4283 | No |
| 684 | PEBP4 | PEBP4 | 3754 | 0.602 | 0.4284 | No |
| 685 | GPC2 | GPC2 | 3780 | 0.602 | 0.4275 | No |
| 686 | GRIP2 | GRIP2 | 3782 | 0.602 | 0.4278 | No |
| 687 | KLRG1 | KLRG1 | 3783 | 0.602 | 0.4282 | No |
| 688 | CHRNA7 | CHRNA7 | 3785 | 0.602 | 0.4285 | No |
| 689 | CHST5 | CHST5 | 3787 | 0.602 | 0.4288 | No |
| 690 | GSX2 | GSX2 | 3805 | 0.602 | 0.4283 | No |
| 691 | AZGP1 | AZGP1 | 3807 | 0.602 | 0.4287 | No |
| 692 | WEE2 | WEE2 | 3809 | 0.602 | 0.4290 | No |
| 693 | PAQR6 | PAQR6 | 3821 | 0.602 | 0.4288 | No |
| 694 | SLC26A1 | SLC26A1 | 3838 | 0.602 | 0.4283 | No |
| 695 | KIF27 | KIF27 | 3857 | 0.602 | 0.4278 | No |
| 696 | ANKRD23 | ANKRD23 | 3873 | 0.602 | 0.4274 | No |
| 697 | PROK1 | PROK1 | 3878 | 0.602 | 0.4276 | No |
| 698 | TTLL9 | TTLL9 | 3893 | 0.602 | 0.4272 | No |
| 699 | GAL3ST4 | GAL3ST4 | 3896 | 0.602 | 0.4275 | No |
| 700 | CERS1 | CERS1 | 3985 | 0.602 | 0.4234 | No |
| 701 | USP11 | USP11 | 4167 | 0.601 | 0.4146 | No |
| 702 | GK5 | GK5 | 4168 | 0.601 | 0.4150 | No |
| 703 | INTS10 | INTS10 | 4179 | 0.600 | 0.4148 | No |
| 704 | ZBTB33 | ZBTB33 | 4183 | 0.599 | 0.4150 | No |
| 705 | VANGL2 | VANGL2 | 4188 | 0.598 | 0.4152 | No |
| 706 | METTL8 | METTL8 | 4195 | 0.595 | 0.4153 | No |
| 707 | DIP2B | DIP2B | 4196 | 0.595 | 0.4156 | No |
| 708 | HYDIN | HYDIN | 4201 | 0.594 | 0.4158 | No |
| 709 | GDF9 | GDF9 | 4204 | 0.594 | 0.4160 | No |
| 710 | FCHSD2 | FCHSD2 | 4205 | 0.593 | 0.4164 | No |
| 711 | KATNAL2 | KATNAL2 | 4208 | 0.592 | 0.4167 | No |
| 712 | TUBA1A | TUBA1A | 4210 | 0.592 | 0.4170 | No |
| 713 | VHL | VHL | 4215 | 0.591 | 0.4171 | No |
| 714 | NPNT | NPNT | 4219 | 0.589 | 0.4173 | No |
| 715 | TUBE1 | TUBE1 | 4221 | 0.589 | 0.4176 | No |
| 716 | CASKIN1 | CASKIN1 | 4222 | 0.589 | 0.4180 | No |
| 717 | DENND6B | DENND6B | 4234 | 0.585 | 0.4178 | No |
| 718 | N4BP2 | N4BP2 | 4238 | 0.585 | 0.4180 | No |
| 719 | PPM1K | PPM1K | 4240 | 0.584 | 0.4183 | No |
| 720 | KLHL25 | KLHL25 | 4243 | 0.584 | 0.4186 | No |
| 721 | HECTD4 | HECTD4 | 4244 | 0.583 | 0.4189 | No |
| 722 | CLIP3 | CLIP3 | 4248 | 0.582 | 0.4191 | No |
| 723 | RNF122 | RNF122 | 4250 | 0.581 | 0.4194 | No |
| 724 | RORC | RORC | 4253 | 0.581 | 0.4197 | No |
| 725 | SMARCD1 | SMARCD1 | 4265 | 0.578 | 0.4195 | No |
| 726 | THRA | THRA | 4266 | 0.578 | 0.4198 | No |
| 727 | CDH22 | CDH22 | 4274 | 0.575 | 0.4198 | No |
| 728 | MDGA2 | MDGA2 | 4278 | 0.574 | 0.4200 | No |
| 729 | MAP3K1 | MAP3K1 | 4285 | 0.571 | 0.4201 | No |
| 730 | CDH10 | CDH10 | 4292 | 0.570 | 0.4201 | No |
| 731 | MAPK8IP1 | MAPK8IP1 | 4305 | 0.565 | 0.4198 | No |
| 732 | ARNT2 | ARNT2 | 4308 | 0.565 | 0.4201 | No |
| 733 | TRIL | TRIL | 4322 | 0.561 | 0.4198 | No |
| 734 | PBX4 | PBX4 | 4324 | 0.561 | 0.4201 | No |
| 735 | CCDC30 | CCDC30 | 4326 | 0.560 | 0.4204 | No |
| 736 | ARHGEF11 | ARHGEF11 | 4327 | 0.560 | 0.4207 | No |
| 737 | USP49 | USP49 | 4330 | 0.558 | 0.4209 | No |
| 738 | SEMA4A | SEMA4A | 4334 | 0.558 | 0.4211 | No |
| 739 | ZKSCAN8 | ZKSCAN8 | 4335 | 0.558 | 0.4215 | No |
| 740 | TRAF6 | TRAF6 | 4341 | 0.557 | 0.4215 | No |
| 741 | CACHD1 | CACHD1 | 4347 | 0.554 | 0.4216 | No |
| 742 | VSIG10L | VSIG10L | 4350 | 0.553 | 0.4219 | No |
| 743 | YBEY | YBEY | 4354 | 0.552 | 0.4220 | No |
| 744 | TTC7B | TTC7B | 4356 | 0.551 | 0.4223 | No |
| 745 | CRY2 | CRY2 | 4359 | 0.551 | 0.4226 | No |
| 746 | EID2B | EID2B | 4368 | 0.550 | 0.4225 | No |
| 747 | FAM107B | FAM107B | 4373 | 0.549 | 0.4226 | No |
| 748 | RIC3 | RIC3 | 4385 | 0.545 | 0.4224 | No |
| 749 | ASB13 | ASB13 | 4387 | 0.544 | 0.4227 | No |
| 750 | BRWD3 | BRWD3 | 4392 | 0.542 | 0.4228 | No |
| 751 | KIF21B | KIF21B | 4393 | 0.542 | 0.4231 | No |
| 752 | SLC13A3 | SLC13A3 | 4395 | 0.541 | 0.4234 | No |
| 753 | CRTC1 | CRTC1 | 4400 | 0.538 | 0.4235 | No |
| 754 | JRK | JRK | 4414 | 0.535 | 0.4232 | No |
| 755 | RAP1GAP | RAP1GAP | 4415 | 0.535 | 0.4235 | No |
| 756 | KDM3B | KDM3B | 4417 | 0.534 | 0.4238 | No |
| 757 | UNK | UNK | 4418 | 0.534 | 0.4241 | No |
| 758 | CRMP1 | CRMP1 | 4419 | 0.534 | 0.4245 | No |
| 759 | GRID1 | GRID1 | 4425 | 0.531 | 0.4245 | No |
| 760 | TATDN3 | TATDN3 | 4427 | 0.531 | 0.4248 | No |
| 761 | CDKL3 | CDKL3 | 4428 | 0.531 | 0.4251 | No |
| 762 | RTN2 | RTN2 | 4430 | 0.530 | 0.4254 | No |
| 763 | CSRNP2 | CSRNP2 | 4431 | 0.530 | 0.4257 | No |
| 764 | WDR47 | WDR47 | 4438 | 0.529 | 0.4257 | No |
| 765 | PYGO1 | PYGO1 | 4444 | 0.529 | 0.4258 | No |
| 766 | PELI1 | PELI1 | 4446 | 0.528 | 0.4261 | No |
| 767 | ZIK1 | ZIK1 | 4458 | 0.526 | 0.4258 | No |
| 768 | VPS37D | VPS37D | 4459 | 0.526 | 0.4262 | No |
| 769 | APBB1 | APBB1 | 4461 | 0.526 | 0.4264 | No |
| 770 | CELSR2 | CELSR2 | 4469 | 0.523 | 0.4264 | No |
| 771 | H1FX | H1FX | 4479 | 0.521 | 0.4263 | No |
| 772 | FRY | FRY | 4487 | 0.519 | 0.4262 | No |
| 773 | GOLGA1 | GOLGA1 | 4490 | 0.519 | 0.4264 | No |
| 774 | DTNB | DTNB | 4499 | 0.517 | 0.4264 | No |
| 775 | MXI1 | MXI1 | 4512 | 0.512 | 0.4261 | No |
| 776 | PPP1R12B | PPP1R12B | 4514 | 0.512 | 0.4263 | No |
| 777 | SLC36A4 | SLC36A4 | 4525 | 0.508 | 0.4261 | No |
| 778 | GADD45G | GADD45G | 4526 | 0.508 | 0.4264 | No |
| 779 | LMTK3 | LMTK3 | 4531 | 0.507 | 0.4265 | No |
| 780 | HRK | HRK | 4534 | 0.505 | 0.4267 | No |
| 781 | TIA1 | TIA1 | 4537 | 0.504 | 0.4269 | No |
| 782 | MTHFD2L | MTHFD2L | 4543 | 0.504 | 0.4270 | No |
| 783 | SHPRH | SHPRH | 4549 | 0.502 | 0.4271 | No |
| 784 | ELMOD3 | ELMOD3 | 4552 | 0.501 | 0.4273 | No |
| 785 | FBRSL1 | FBRSL1 | 4553 | 0.501 | 0.4276 | No |
| 786 | GMIP | GMIP | 4564 | 0.499 | 0.4274 | No |
| 787 | PPM1D | PPM1D | 4569 | 0.498 | 0.4275 | No |
| 788 | PIK3R3 | PIK3R3 | 4571 | 0.498 | 0.4277 | No |
| 789 | SLC35A5 | SLC35A5 | 4577 | 0.496 | 0.4278 | No |
| 790 | XYLT1 | XYLT1 | 4580 | 0.496 | 0.4280 | No |
| 791 | MMP16 | MMP16 | 4597 | 0.491 | 0.4275 | No |
| 792 | RBAK | RBAK | 4599 | 0.490 | 0.4277 | No |
| 793 | FIGN | FIGN | 4600 | 0.490 | 0.4280 | No |
| 794 | ANKRD16 | ANKRD16 | 4602 | 0.490 | 0.4282 | No |
| 795 | CREBZF | CREBZF | 4607 | 0.489 | 0.4283 | No |
| 796 | ZFP2 | ZFP2 | 4610 | 0.488 | 0.4285 | No |
| 797 | AGO4 | AGO4 | 4613 | 0.487 | 0.4287 | No |
| 798 | RMND5B | RMND5B | 4623 | 0.485 | 0.4286 | No |
| 799 | NRXN2 | NRXN2 | 4628 | 0.483 | 0.4287 | No |
| 800 | ACOT6 | ACOT6 | 4633 | 0.482 | 0.4288 | No |
| 801 | KMT2C | KMT2C | 4637 | 0.481 | 0.4289 | No |
| 802 | TRIM23 | TRIM23 | 4650 | 0.478 | 0.4286 | No |
| 803 | ADCY1 | ADCY1 | 4653 | 0.477 | 0.4288 | No |
| 804 | USP3 | USP3 | 4660 | 0.477 | 0.4288 | No |
| 805 | DNAJC28 | DNAJC28 | 4662 | 0.476 | 0.4290 | No |
| 806 | TSPAN7 | TSPAN7 | 4665 | 0.475 | 0.4292 | No |
| 807 | RFXANK | RFXANK | 4668 | 0.475 | 0.4294 | No |
| 808 | SRR | SRR | 4688 | 0.473 | 0.4287 | No |
| 809 | ZMYM2 | ZMYM2 | 4694 | 0.472 | 0.4287 | No |
| 810 | SS18L1 | SS18L1 | 4695 | 0.472 | 0.4290 | No |
| 811 | PAN2 | PAN2 | 4696 | 0.472 | 0.4293 | No |
| 812 | KANK3 | KANK3 | 4700 | 0.471 | 0.4294 | No |
| 813 | FAM171A2 | FAM171A2 | 4701 | 0.471 | 0.4297 | No |
| 814 | ACAP3 | ACAP3 | 4706 | 0.470 | 0.4298 | No |
| 815 | HIC2 | HIC2 | 4707 | 0.470 | 0.4301 | No |
| 816 | MT3 | MT3 | 4714 | 0.469 | 0.4301 | No |
| 817 | CACNB4 | CACNB4 | 4725 | 0.467 | 0.4299 | No |
| 818 | APC | APC | 4727 | 0.467 | 0.4301 | No |
| 819 | KDM4B | KDM4B | 4731 | 0.466 | 0.4302 | No |
| 820 | DUSP28 | DUSP28 | 4740 | 0.464 | 0.4301 | No |
| 821 | LINGO1 | LINGO1 | 4743 | 0.464 | 0.4303 | No |
| 822 | NAV1 | NAV1 | 4744 | 0.463 | 0.4306 | No |
| 823 | SYNGAP1 | SYNGAP1 | 4745 | 0.463 | 0.4308 | No |
| 824 | CCDC13 | CCDC13 | 4756 | 0.461 | 0.4306 | No |
| 825 | SOX2 | SOX2 | 4761 | 0.460 | 0.4307 | No |
| 826 | CAMSAP3 | CAMSAP3 | 4762 | 0.460 | 0.4310 | No |
| 827 | SSBP3 | SSBP3 | 4770 | 0.459 | 0.4309 | No |
| 828 | LRP4 | LRP4 | 4771 | 0.459 | 0.4312 | No |
| 829 | ATAD2B | ATAD2B | 4773 | 0.459 | 0.4314 | No |
| 830 | CEP97 | CEP97 | 4775 | 0.458 | 0.4316 | No |
| 831 | CHD7 | CHD7 | 4776 | 0.458 | 0.4319 | No |
| 832 | CLIP2 | CLIP2 | 4783 | 0.458 | 0.4319 | No |
| 833 | SEPHS1 | SEPHS1 | 4792 | 0.456 | 0.4318 | No |
| 834 | PFKFB1 | PFKFB1 | 4796 | 0.456 | 0.4319 | No |
| 835 | TAF4 | TAF4 | 4798 | 0.456 | 0.4321 | No |
| 836 | MARCKS | MARCKS | 4801 | 0.455 | 0.4323 | No |
| 837 | YEATS2 | YEATS2 | 4806 | 0.454 | 0.4324 | No |
| 838 | MVB12B | MVB12B | 4810 | 0.453 | 0.4325 | No |
| 839 | PCCA | PCCA | 4811 | 0.453 | 0.4328 | No |
| 840 | GANC | GANC | 4817 | 0.451 | 0.4328 | No |
| 841 | SATB2 | SATB2 | 4820 | 0.451 | 0.4330 | No |
| 842 | ZBTB26 | ZBTB26 | 4821 | 0.451 | 0.4332 | No |
| 843 | KCNN1 | KCNN1 | 4825 | 0.450 | 0.4334 | No |
| 844 | ST6GAL1 | ST6GAL1 | 4828 | 0.449 | 0.4335 | No |
| 845 | METTL9 | METTL9 | 4829 | 0.449 | 0.4338 | No |
| 846 | GABBR1 | GABBR1 | 4840 | 0.447 | 0.4336 | No |
| 847 | RGS12 | RGS12 | 4841 | 0.447 | 0.4338 | No |
| 848 | LRFN1 | LRFN1 | 4844 | 0.447 | 0.4340 | No |
| 849 | TTYH2 | TTYH2 | 4847 | 0.446 | 0.4342 | No |
| 850 | NPVF | NPVF | 4853 | 0.445 | 0.4342 | No |
| 851 | CTC1 | CTC1 | 4855 | 0.444 | 0.4344 | No |
| 852 | PACSIN1 | PACSIN1 | 4861 | 0.443 | 0.4344 | No |
| 853 | PCED1A | PCED1A | 4867 | 0.443 | 0.4345 | No |
| 854 | DEPDC5 | DEPDC5 | 4872 | 0.442 | 0.4345 | No |
| 855 | SOX9 | SOX9 | 4879 | 0.441 | 0.4345 | No |
| 856 | ACVR1B | ACVR1B | 4883 | 0.439 | 0.4346 | No |
| 857 | KBTBD6 | KBTBD6 | 4886 | 0.439 | 0.4348 | No |
| 858 | OPTC | OPTC | 4893 | 0.438 | 0.4347 | No |
| 859 | FAM19A5 | FAM19A5 | 4896 | 0.438 | 0.4349 | No |
| 860 | SEC61A2 | SEC61A2 | 4901 | 0.436 | 0.4350 | No |
| 861 | MSI2 | MSI2 | 4908 | 0.435 | 0.4349 | No |
| 862 | ZBTB5 | ZBTB5 | 4909 | 0.435 | 0.4352 | No |
| 863 | SETD4 | SETD4 | 4910 | 0.435 | 0.4355 | No |
| 864 | WSB1 | WSB1 | 4911 | 0.435 | 0.4357 | No |
| 865 | TBCCD1 | TBCCD1 | 4914 | 0.434 | 0.4359 | No |
| 866 | WDR83 | WDR83 | 4920 | 0.433 | 0.4359 | No |
| 867 | MARK1 | MARK1 | 4921 | 0.433 | 0.4362 | No |
| 868 | PRDM15 | PRDM15 | 4924 | 0.432 | 0.4363 | No |
| 869 | MORN4 | MORN4 | 4928 | 0.432 | 0.4364 | No |
| 870 | CDH20 | CDH20 | 4931 | 0.431 | 0.4366 | No |
| 871 | DCHS1 | DCHS1 | 4936 | 0.431 | 0.4366 | No |
| 872 | LMF1 | LMF1 | 4938 | 0.431 | 0.4369 | No |
| 873 | CLCN2 | CLCN2 | 4949 | 0.430 | 0.4366 | No |
| 874 | INO80B | INO80B | 4969 | 0.426 | 0.4359 | No |
| 875 | SPICE1 | SPICE1 | 4988 | 0.422 | 0.4353 | No |
| 876 | ELDR | ELDR | 4991 | 0.422 | 0.4354 | No |
| 877 | ACBD5 | ACBD5 | 4993 | 0.422 | 0.4356 | No |
| 878 | PCDHGA4 | PCDHGA4 | 4997 | 0.421 | 0.4357 | No |
| 879 | JPH4 | JPH4 | 4998 | 0.421 | 0.4360 | No |
| 880 | CCDC136 | CCDC136 | 5003 | 0.420 | 0.4360 | No |
| 881 | GDPD1 | GDPD1 | 5004 | 0.420 | 0.4363 | No |
| 882 | GPHN | GPHN | 5006 | 0.420 | 0.4365 | No |
| 883 | PDGFRA | PDGFRA | 5008 | 0.420 | 0.4367 | No |
| 884 | PLA2G6 | PLA2G6 | 5010 | 0.419 | 0.4369 | No |
| 885 | POLR2F | POLR2F | 5017 | 0.419 | 0.4369 | No |
| 886 | TMEFF1 | TMEFF1 | 5020 | 0.418 | 0.4370 | No |
| 887 | PRELP | PRELP | 5027 | 0.417 | 0.4370 | No |
| 888 | SPAST | SPAST | 5029 | 0.417 | 0.4372 | No |
| 889 | KLHDC3 | KLHDC3 | 5032 | 0.416 | 0.4373 | No |
| 890 | UBXN2B | UBXN2B | 5034 | 0.416 | 0.4375 | No |
| 891 | DNAL1 | DNAL1 | 5036 | 0.415 | 0.4377 | No |
| 892 | SDC3 | SDC3 | 5038 | 0.415 | 0.4379 | No |
| 893 | KSR1 | KSR1 | 5052 | 0.413 | 0.4375 | No |
| 894 | NOTCH1 | NOTCH1 | 5062 | 0.412 | 0.4373 | No |
| 895 | NLGN2 | NLGN2 | 5067 | 0.412 | 0.4374 | No |
| 896 | EPB41L5 | EPB41L5 | 5069 | 0.411 | 0.4376 | No |
| 897 | ARID2 | ARID2 | 5076 | 0.410 | 0.4375 | No |
| 898 | ATXN7L2 | ATXN7L2 | 5082 | 0.409 | 0.4375 | No |
| 899 | LSM14B | LSM14B | 5083 | 0.408 | 0.4377 | No |
| 900 | GTF2I | GTF2I | 5088 | 0.407 | 0.4378 | No |
| 901 | PPARGC1A | PPARGC1A | 5094 | 0.406 | 0.4378 | No |
| 902 | ZMYND8 | ZMYND8 | 5096 | 0.406 | 0.4380 | No |
| 903 | DPYSL2 | DPYSL2 | 5098 | 0.406 | 0.4382 | No |
| 904 | KIF1B | KIF1B | 5101 | 0.405 | 0.4383 | No |
| 905 | GNG8 | GNG8 | 5102 | 0.405 | 0.4386 | No |
| 906 | RIIAD1 | RIIAD1 | 5112 | 0.404 | 0.4384 | No |
| 907 | POLR3GL | POLR3GL | 5120 | 0.402 | 0.4383 | No |
| 908 | RASGRP2 | RASGRP2 | 5124 | 0.402 | 0.4383 | No |
| 909 | SHISA4 | SHISA4 | 5125 | 0.401 | 0.4386 | No |
| 910 | TMOD2 | TMOD2 | 5141 | 0.400 | 0.4381 | No |
| 911 | LMO2 | LMO2 | 5143 | 0.400 | 0.4383 | No |
| 912 | PFKM | PFKM | 5149 | 0.399 | 0.4383 | No |
| 913 | ZBTB37 | ZBTB37 | 5150 | 0.399 | 0.4385 | No |
| 914 | DNALI1 | DNALI1 | 5151 | 0.399 | 0.4387 | No |
| 915 | TMEM38A | TMEM38A | 5155 | 0.398 | 0.4388 | No |
| 916 | NPAS3 | NPAS3 | 5163 | 0.397 | 0.4387 | No |
| 917 | GRAMD1A | GRAMD1A | 5167 | 0.396 | 0.4388 | No |
| 918 | CAMK2N2 | CAMK2N2 | 5169 | 0.396 | 0.4390 | No |
| 919 | ATAT1 | ATAT1 | 5178 | 0.394 | 0.4388 | No |
| 920 | BBS2 | BBS2 | 5189 | 0.393 | 0.4386 | No |
| 921 | SP4 | SP4 | 5203 | 0.390 | 0.4381 | No |
| 922 | TMEM198B | TMEM198B | 5205 | 0.390 | 0.4383 | No |
| 923 | CBX8 | CBX8 | 5207 | 0.389 | 0.4385 | No |
| 924 | MGAT3 | MGAT3 | 5208 | 0.389 | 0.4388 | No |
| 925 | TTLL1 | TTLL1 | 5218 | 0.387 | 0.4385 | No |
| 926 | APC2 | APC2 | 5221 | 0.387 | 0.4387 | No |
| 927 | CCT6B | CCT6B | 5224 | 0.387 | 0.4388 | No |
| 928 | BRD7 | BRD7 | 5236 | 0.386 | 0.4385 | No |
| 929 | ADAM22 | ADAM22 | 5263 | 0.382 | 0.4374 | No |
| 930 | SETDB1 | SETDB1 | 5267 | 0.381 | 0.4375 | No |
| 931 | SENP7 | SENP7 | 5280 | 0.379 | 0.4371 | No |
| 932 | KLHDC8B | KLHDC8B | 5281 | 0.379 | 0.4373 | No |
| 933 | TET1 | TET1 | 5287 | 0.379 | 0.4373 | No |
| 934 | MTSS1 | MTSS1 | 5291 | 0.378 | 0.4374 | No |
| 935 | SALL1 | SALL1 | 5296 | 0.378 | 0.4374 | No |
| 936 | DDX25 | DDX25 | 5298 | 0.378 | 0.4376 | No |
| 937 | KDM2B | KDM2B | 5299 | 0.378 | 0.4378 | No |
| 938 | FAM120C | FAM120C | 5301 | 0.378 | 0.4380 | No |
| 939 | PIGX | PIGX | 5307 | 0.377 | 0.4380 | No |
| 940 | FAM168A | FAM168A | 5308 | 0.377 | 0.4382 | No |
| 941 | PNN | PNN | 5310 | 0.376 | 0.4384 | No |
| 942 | CFAP74 | CFAP74 | 5314 | 0.376 | 0.4385 | No |
| 943 | TSPYL4 | TSPYL4 | 5323 | 0.373 | 0.4383 | No |
| 944 | NAA60 | NAA60 | 5332 | 0.373 | 0.4381 | No |
| 945 | BRSK1 | BRSK1 | 5338 | 0.372 | 0.4381 | No |
| 946 | FBXO11 | FBXO11 | 5352 | 0.370 | 0.4377 | No |
| 947 | PATZ1 | PATZ1 | 5362 | 0.369 | 0.4374 | No |
| 948 | RUNDC3A | RUNDC3A | 5364 | 0.369 | 0.4376 | No |
| 949 | KLHL24 | KLHL24 | 5365 | 0.369 | 0.4378 | No |
| 950 | SFI1 | SFI1 | 5371 | 0.368 | 0.4378 | No |
| 951 | ATP6V0A1 | ATP6V0A1 | 5374 | 0.367 | 0.4379 | No |
| 952 | SAP25 | SAP25 | 5384 | 0.365 | 0.4377 | No |
| 953 | LIN52 | LIN52 | 5389 | 0.365 | 0.4377 | No |
| 954 | CDKN1B | CDKN1B | 5400 | 0.364 | 0.4374 | No |
| 955 | TTBK2 | TTBK2 | 5405 | 0.363 | 0.4374 | No |
| 956 | RCOR2 | RCOR2 | 5406 | 0.363 | 0.4377 | No |
| 957 | ADGRL1 | ADGRL1 | 5408 | 0.363 | 0.4378 | No |
| 958 | ACAD10 | ACAD10 | 5416 | 0.362 | 0.4377 | No |
| 959 | CHRDL1 | CHRDL1 | 5422 | 0.361 | 0.4377 | No |
| 960 | YPEL3 | YPEL3 | 5427 | 0.360 | 0.4377 | No |
| 961 | PEX5 | PEX5 | 5430 | 0.359 | 0.4378 | No |
| 962 | AKAP7 | AKAP7 | 5433 | 0.359 | 0.4379 | No |
| 963 | TM6SF2 | TM6SF2 | 5434 | 0.359 | 0.4381 | No |
| 964 | GRB14 | GRB14 | 5437 | 0.359 | 0.4383 | No |
| 965 | VRK3 | VRK3 | 5438 | 0.359 | 0.4385 | No |
| 966 | SHISA8 | SHISA8 | 5440 | 0.359 | 0.4386 | No |
| 967 | PALD1 | PALD1 | 5444 | 0.358 | 0.4387 | No |
| 968 | SYT11 | SYT11 | 5449 | 0.357 | 0.4387 | No |
| 969 | CAMK2B | CAMK2B | 5451 | 0.357 | 0.4389 | No |
| 970 | SRCIN1 | SRCIN1 | 5454 | 0.356 | 0.4390 | No |
| 971 | TMEM179 | TMEM179 | 5456 | 0.356 | 0.4392 | No |
| 972 | SALL3 | SALL3 | 5460 | 0.356 | 0.4392 | No |
| 973 | IFT140 | IFT140 | 5466 | 0.355 | 0.4392 | No |
| 974 | TLE1 | TLE1 | 5468 | 0.355 | 0.4394 | No |
| 975 | IQCE | IQCE | 5476 | 0.354 | 0.4392 | No |
| 976 | GDI1 | GDI1 | 5480 | 0.354 | 0.4393 | No |
| 977 | DCAF17 | DCAF17 | 5482 | 0.353 | 0.4394 | No |
| 978 | RBM5 | RBM5 | 5489 | 0.351 | 0.4394 | No |
| 979 | FAM198A | FAM198A | 5491 | 0.351 | 0.4395 | No |
| 980 | FBXO36 | FBXO36 | 5498 | 0.350 | 0.4394 | No |
| 981 | ZFP62 | ZFP62 | 5499 | 0.350 | 0.4396 | No |
| 982 | BTF3L4 | BTF3L4 | 5509 | 0.348 | 0.4394 | No |
| 983 | SOX12 | SOX12 | 5515 | 0.347 | 0.4394 | No |
| 984 | SRGAP2 | SRGAP2 | 5516 | 0.347 | 0.4396 | No |
| 985 | ANKRD46 | ANKRD46 | 5522 | 0.346 | 0.4395 | No |
| 986 | PPOX | PPOX | 5527 | 0.345 | 0.4395 | No |
| 987 | TRIB2 | TRIB2 | 5529 | 0.344 | 0.4397 | No |
| 988 | LDLRAD3 | LDLRAD3 | 5533 | 0.344 | 0.4398 | No |
| 989 | FAM84A | FAM84A | 5535 | 0.344 | 0.4399 | No |
| 990 | TADA1 | TADA1 | 5539 | 0.344 | 0.4400 | No |
| 991 | TMEM206 | TMEM206 | 5545 | 0.343 | 0.4399 | No |
| 992 | KDM3A | KDM3A | 5553 | 0.341 | 0.4398 | No |
| 993 | MBTD1 | MBTD1 | 5564 | 0.340 | 0.4395 | No |
| 994 | RAB3A | RAB3A | 5567 | 0.339 | 0.4396 | No |
| 995 | TTC14 | TTC14 | 5574 | 0.339 | 0.4395 | No |
| 996 | RORA | RORA | 5575 | 0.338 | 0.4397 | No |
| 997 | SOX4 | SOX4 | 5577 | 0.338 | 0.4399 | No |
| 998 | COX20 | COX20 | 5579 | 0.338 | 0.4400 | No |
| 999 | ARHGEF7 | ARHGEF7 | 5580 | 0.338 | 0.4402 | No |
| 1000 | WDR19 | WDR19 | 5582 | 0.338 | 0.4404 | No |
| 1001 | PEX1 | PEX1 | 5587 | 0.337 | 0.4404 | No |
| 1002 | NECAB2 | NECAB2 | 5588 | 0.337 | 0.4406 | No |
| 1003 | TAB1 | TAB1 | 5591 | 0.337 | 0.4407 | No |
| 1004 | NICN1 | NICN1 | 5598 | 0.336 | 0.4406 | No |
| 1005 | FYN | FYN | 5599 | 0.336 | 0.4408 | No |
| 1006 | CAMSAP2 | CAMSAP2 | 5601 | 0.336 | 0.4409 | No |
| 1007 | TNFRSF21 | TNFRSF21 | 5604 | 0.335 | 0.4410 | No |
| 1008 | DDX17 | DDX17 | 5608 | 0.334 | 0.4411 | No |
| 1009 | MLXIP | MLXIP | 5623 | 0.332 | 0.4406 | No |
| 1010 | CLK1 | CLK1 | 5625 | 0.332 | 0.4407 | No |
| 1011 | ATP6V0E2 | ATP6V0E2 | 5627 | 0.332 | 0.4409 | No |
| 1012 | PIAS2 | PIAS2 | 5629 | 0.332 | 0.4410 | No |
| 1013 | ONECUT2 | ONECUT2 | 5637 | 0.331 | 0.4409 | No |
| 1014 | SGSM1 | SGSM1 | 5638 | 0.331 | 0.4411 | No |
| 1015 | ELP4 | ELP4 | 5648 | 0.329 | 0.4408 | No |
| 1016 | RAB11B | RAB11B | 5649 | 0.329 | 0.4410 | No |
| 1017 | BAZ2B | BAZ2B | 5650 | 0.329 | 0.4412 | No |
| 1018 | ANP32A | ANP32A | 5651 | 0.329 | 0.4414 | No |
| 1019 | PHF14 | PHF14 | 5655 | 0.328 | 0.4415 | No |
| 1020 | CHFR | CHFR | 5667 | 0.327 | 0.4411 | No |
| 1021 | MYEF2 | MYEF2 | 5670 | 0.326 | 0.4412 | No |
| 1022 | TDRKH | TDRKH | 5675 | 0.326 | 0.4412 | No |
| 1023 | CEP72 | CEP72 | 5676 | 0.325 | 0.4414 | No |
| 1024 | TOP2B | TOP2B | 5705 | 0.321 | 0.4402 | No |
| 1025 | POGK | POGK | 5706 | 0.321 | 0.4404 | No |
| 1026 | RIMS4 | RIMS4 | 5707 | 0.320 | 0.4406 | No |
| 1027 | MYO6 | MYO6 | 5711 | 0.320 | 0.4406 | No |
| 1028 | PTPRS | PTPRS | 5723 | 0.318 | 0.4403 | No |
| 1029 | DGCR6 | DGCR6 | 5735 | 0.317 | 0.4399 | No |
| 1030 | NPDC1 | NPDC1 | 5739 | 0.317 | 0.4399 | No |
| 1031 | CLK4 | CLK4 | 5741 | 0.317 | 0.4401 | No |
| 1032 | CRAMP1L | CRAMP1L | 5742 | 0.317 | 0.4403 | No |
| 1033 | HPS4 | HPS4 | 5746 | 0.316 | 0.4403 | No |
| 1034 | EEF2K | EEF2K | 5754 | 0.315 | 0.4402 | No |
| 1035 | EBF4 | EBF4 | 5761 | 0.314 | 0.4400 | No |
| 1036 | TNFAIP6 | TNFAIP6 | 5763 | 0.314 | 0.4402 | No |
| 1037 | MTTP | MTTP | 5764 | 0.314 | 0.4404 | No |
| 1038 | STRIP2 | STRIP2 | 5781 | 0.312 | 0.4398 | No |
| 1039 | PTPRA | PTPRA | 5785 | 0.312 | 0.4398 | No |
| 1040 | CPSF6 | CPSF6 | 5803 | 0.310 | 0.4391 | No |
| 1041 | DIP2A | DIP2A | 5815 | 0.309 | 0.4388 | No |
| 1042 | PGPEP1 | PGPEP1 | 5817 | 0.309 | 0.4389 | No |
| 1043 | SNX32 | SNX32 | 5818 | 0.309 | 0.4391 | No |
| 1044 | RBM33 | RBM33 | 5822 | 0.309 | 0.4391 | No |
| 1045 | PAXBP1 | PAXBP1 | 5824 | 0.309 | 0.4393 | No |
| 1046 | CASK | CASK | 5839 | 0.306 | 0.4387 | No |
| 1047 | ATG16L2 | ATG16L2 | 5847 | 0.305 | 0.4386 | No |
| 1048 | ARL15 | ARL15 | 5852 | 0.305 | 0.4385 | No |
| 1049 | TNRC6C | TNRC6C | 5869 | 0.304 | 0.4379 | No |
| 1050 | LCORL | LCORL | 5876 | 0.303 | 0.4378 | No |
| 1051 | DVL3 | DVL3 | 5877 | 0.303 | 0.4380 | No |
| 1052 | MARK4 | MARK4 | 5889 | 0.301 | 0.4376 | No |
| 1053 | LMBR1L | LMBR1L | 5890 | 0.301 | 0.4378 | No |
| 1054 | HIGD2A | HIGD2A | 5901 | 0.299 | 0.4375 | No |
| 1055 | HSD17B11 | HSD17B11 | 5904 | 0.299 | 0.4376 | No |
| 1056 | ATG4D | ATG4D | 5906 | 0.298 | 0.4377 | No |
| 1057 | PJA1 | PJA1 | 5911 | 0.297 | 0.4377 | No |
| 1058 | PHF21A | PHF21A | 5930 | 0.295 | 0.4369 | No |
| 1059 | CLK2 | CLK2 | 5932 | 0.295 | 0.4371 | No |
| 1060 | RFX7 | RFX7 | 5935 | 0.295 | 0.4371 | No |
| 1061 | PTOV1 | PTOV1 | 5936 | 0.295 | 0.4373 | No |
| 1062 | RIC8B | RIC8B | 5950 | 0.293 | 0.4368 | No |
| 1063 | FAM219B | FAM219B | 5957 | 0.292 | 0.4367 | No |
| 1064 | DLGAP1 | DLGAP1 | 5963 | 0.292 | 0.4366 | No |
| 1065 | FBXO46 | FBXO46 | 5966 | 0.291 | 0.4367 | No |
| 1066 | HES6 | HES6 | 5983 | 0.288 | 0.4361 | No |
| 1067 | METTL21A | METTL21A | 5988 | 0.288 | 0.4361 | No |
| 1068 | MARS2 | MARS2 | 5989 | 0.287 | 0.4362 | No |
| 1069 | KLHL32 | KLHL32 | 5994 | 0.287 | 0.4362 | No |
| 1070 | CCNL2 | CCNL2 | 5995 | 0.287 | 0.4364 | No |
| 1071 | DOT1L | DOT1L | 5998 | 0.287 | 0.4364 | No |
| 1072 | KANSL1 | KANSL1 | 6000 | 0.287 | 0.4366 | No |
| 1073 | TMEM116 | TMEM116 | 6002 | 0.286 | 0.4367 | No |
| 1074 | JAM3 | JAM3 | 6015 | 0.285 | 0.4363 | No |
| 1075 | MCCC1 | MCCC1 | 6024 | 0.284 | 0.4360 | No |
| 1076 | TRIM36 | TRIM36 | 6029 | 0.283 | 0.4360 | No |
| 1077 | ZMYM3 | ZMYM3 | 6031 | 0.283 | 0.4361 | No |
| 1078 | NCAM1 | NCAM1 | 6035 | 0.282 | 0.4361 | No |
| 1079 | MAP3K12 | MAP3K12 | 6039 | 0.282 | 0.4362 | No |
| 1080 | PHLPP1 | PHLPP1 | 6044 | 0.281 | 0.4361 | No |
| 1081 | KLHL23 | KLHL23 | 6045 | 0.281 | 0.4363 | No |
| 1082 | FIP1L1 | FIP1L1 | 6046 | 0.281 | 0.4365 | No |
| 1083 | TEKT2 | TEKT2 | 6062 | 0.279 | 0.4359 | No |
| 1084 | CRKL | CRKL | 6064 | 0.279 | 0.4360 | No |
| 1085 | NPR1 | NPR1 | 6067 | 0.278 | 0.4361 | No |
| 1086 | USP54 | USP54 | 6069 | 0.278 | 0.4362 | No |
| 1087 | EFNA3 | EFNA3 | 6072 | 0.278 | 0.4363 | No |
| 1088 | SLC25A12 | SLC25A12 | 6087 | 0.277 | 0.4357 | No |
| 1089 | GABARAPL2 | GABARAPL2 | 6088 | 0.277 | 0.4359 | No |
| 1090 | NCOA1 | NCOA1 | 6089 | 0.277 | 0.4361 | No |
| 1091 | NDUFC2 | NDUFC2 | 6093 | 0.276 | 0.4361 | No |
| 1092 | ZFP30 | ZFP30 | 6097 | 0.276 | 0.4361 | No |
| 1093 | CDS2 | CDS2 | 6101 | 0.275 | 0.4361 | No |
| 1094 | PCSK4 | PCSK4 | 6105 | 0.275 | 0.4361 | No |
| 1095 | PCIF1 | PCIF1 | 6117 | 0.273 | 0.4357 | No |
| 1096 | TRAPPC6A | TRAPPC6A | 6142 | 0.270 | 0.4347 | No |
| 1097 | PLCG1 | PLCG1 | 6167 | 0.267 | 0.4336 | No |
| 1098 | TRIM33 | TRIM33 | 6170 | 0.267 | 0.4337 | No |
| 1099 | SHF | SHF | 6194 | 0.264 | 0.4327 | No |
| 1100 | CNTD1 | CNTD1 | 6202 | 0.263 | 0.4325 | No |
| 1101 | PRPF38A | PRPF38A | 6215 | 0.263 | 0.4320 | No |
| 1102 | MED25 | MED25 | 6223 | 0.262 | 0.4318 | No |
| 1103 | VPS26B | VPS26B | 6225 | 0.262 | 0.4320 | No |
| 1104 | MDM4 | MDM4 | 6226 | 0.262 | 0.4321 | No |
| 1105 | ACTR3B | ACTR3B | 6232 | 0.261 | 0.4320 | No |
| 1106 | SIRT2 | SIRT2 | 6235 | 0.261 | 0.4321 | No |
| 1107 | SIRT3 | SIRT3 | 6240 | 0.260 | 0.4320 | No |
| 1108 | USF1 | USF1 | 6242 | 0.260 | 0.4321 | No |
| 1109 | PNMAL2 | PNMAL2 | 6248 | 0.260 | 0.4320 | No |
| 1110 | IRGQ | IRGQ | 6252 | 0.259 | 0.4321 | No |
| 1111 | TAZ | TAZ | 6254 | 0.258 | 0.4322 | No |
| 1112 | TMCC1 | TMCC1 | 6256 | 0.258 | 0.4323 | No |
| 1113 | MLLT6 | MLLT6 | 6260 | 0.258 | 0.4323 | No |
| 1114 | DEAF1 | DEAF1 | 6263 | 0.258 | 0.4323 | No |
| 1115 | CELF2 | CELF2 | 6266 | 0.257 | 0.4324 | No |
| 1116 | SAT2 | SAT2 | 6268 | 0.257 | 0.4325 | No |
| 1117 | SSBP2 | SSBP2 | 6275 | 0.257 | 0.4323 | No |
| 1118 | KLHL42 | KLHL42 | 6281 | 0.256 | 0.4322 | No |
| 1119 | PHC1 | PHC1 | 6285 | 0.256 | 0.4322 | No |
| 1120 | SPPL2B | SPPL2B | 6286 | 0.255 | 0.4324 | No |
| 1121 | RPAIN | RPAIN | 6290 | 0.255 | 0.4324 | No |
| 1122 | FREM1 | FREM1 | 6291 | 0.255 | 0.4326 | No |
| 1123 | GNAO1 | GNAO1 | 6298 | 0.254 | 0.4324 | No |
| 1124 | EPC2 | EPC2 | 6301 | 0.254 | 0.4325 | No |
| 1125 | CYB5RL | CYB5RL | 6309 | 0.253 | 0.4323 | No |
| 1126 | TTL | TTL | 6310 | 0.253 | 0.4324 | No |
| 1127 | RAB40C | RAB40C | 6318 | 0.252 | 0.4322 | No |
| 1128 | LRRC24 | LRRC24 | 6320 | 0.252 | 0.4323 | No |
| 1129 | FBXL20 | FBXL20 | 6324 | 0.251 | 0.4323 | No |
| 1130 | GNPAT | GNPAT | 6325 | 0.251 | 0.4325 | No |
| 1131 | TBC1D24 | TBC1D24 | 6327 | 0.251 | 0.4326 | No |
| 1132 | RBSN | RBSN | 6340 | 0.249 | 0.4321 | No |
| 1133 | TMEM222 | TMEM222 | 6344 | 0.249 | 0.4321 | No |
| 1134 | STK11IP | STK11IP | 6348 | 0.248 | 0.4321 | No |
| 1135 | SIKE1 | SIKE1 | 6354 | 0.248 | 0.4320 | No |
| 1136 | LRFN4 | LRFN4 | 6355 | 0.248 | 0.4322 | No |
| 1137 | KDM7A | KDM7A | 6356 | 0.247 | 0.4323 | No |
| 1138 | UBOX5 | UBOX5 | 6361 | 0.246 | 0.4323 | No |
| 1139 | DBI | DBI | 6371 | 0.245 | 0.4320 | No |
| 1140 | GPM6B | GPM6B | 6372 | 0.245 | 0.4321 | No |
| 1141 | SNHG20 | SNHG20 | 6375 | 0.245 | 0.4322 | No |
| 1142 | PRDX2 | PRDX2 | 6379 | 0.245 | 0.4322 | No |
| 1143 | FAM131A | FAM131A | 6382 | 0.244 | 0.4322 | No |
| 1144 | HSBP1 | HSBP1 | 6383 | 0.244 | 0.4324 | No |
| 1145 | SNAPIN | SNAPIN | 6397 | 0.242 | 0.4318 | No |
| 1146 | CCDC138 | CCDC138 | 6434 | 0.241 | 0.4302 | No |
| 1147 | ELF2 | ELF2 | 6440 | 0.240 | 0.4301 | No |
| 1148 | RAB39B | RAB39B | 6442 | 0.240 | 0.4302 | No |
| 1149 | ASB8 | ASB8 | 6444 | 0.240 | 0.4303 | No |
| 1150 | KIDINS220 | KIDINS220 | 6449 | 0.239 | 0.4302 | No |
| 1151 | PARP11 | PARP11 | 6474 | 0.237 | 0.4291 | No |
| 1152 | NT5M | NT5M | 6480 | 0.237 | 0.4290 | No |
| 1153 | RNU11 | RNU11 | 6485 | 0.236 | 0.4290 | No |
| 1154 | UBN2 | UBN2 | 6495 | 0.236 | 0.4286 | No |
| 1155 | EIF2AK1 | EIF2AK1 | 6505 | 0.235 | 0.4283 | No |
| 1156 | DMTF1 | DMTF1 | 6514 | 0.234 | 0.4281 | No |
| 1157 | PLEKHA4 | PLEKHA4 | 6519 | 0.233 | 0.4280 | No |
| 1158 | H2AFV | H2AFV | 6530 | 0.233 | 0.4276 | No |
| 1159 | VAMP2 | VAMP2 | 6540 | 0.232 | 0.4273 | No |
| 1160 | DENND5A | DENND5A | 6554 | 0.230 | 0.4268 | No |
| 1161 | NKAIN1 | NKAIN1 | 6558 | 0.230 | 0.4268 | No |
| 1162 | PALM | PALM | 6560 | 0.229 | 0.4269 | No |
| 1163 | CXXC1 | CXXC1 | 6563 | 0.229 | 0.4269 | No |
| 1164 | PKNOX2 | PKNOX2 | 6567 | 0.229 | 0.4269 | No |
| 1165 | RSAD1 | RSAD1 | 6574 | 0.228 | 0.4268 | No |
| 1166 | EPHB3 | EPHB3 | 6576 | 0.228 | 0.4268 | No |
| 1167 | MLYCD | MLYCD | 6577 | 0.227 | 0.4270 | No |
| 1168 | ARID1A | ARID1A | 6582 | 0.227 | 0.4269 | No |
| 1169 | WBP1 | WBP1 | 6587 | 0.226 | 0.4268 | No |
| 1170 | GRM5 | GRM5 | 6592 | 0.226 | 0.4268 | No |
| 1171 | YAF2 | YAF2 | 6594 | 0.225 | 0.4269 | No |
| 1172 | FAM181B | FAM181B | 6608 | 0.224 | 0.4263 | No |
| 1173 | HSDL1 | HSDL1 | 6610 | 0.224 | 0.4264 | No |
| 1174 | SPEN | SPEN | 6617 | 0.224 | 0.4263 | No |
| 1175 | KAT2B | KAT2B | 6622 | 0.224 | 0.4262 | No |
| 1176 | KBTBD3 | KBTBD3 | 6624 | 0.224 | 0.4263 | No |
| 1177 | PTP4A3 | PTP4A3 | 6629 | 0.222 | 0.4262 | No |
| 1178 | PCDHGB2 | PCDHGB2 | 6648 | 0.221 | 0.4254 | No |
| 1179 | ZMYND11 | ZMYND11 | 6666 | 0.219 | 0.4247 | No |
| 1180 | USP24 | USP24 | 6670 | 0.219 | 0.4247 | No |
| 1181 | ARGLU1 | ARGLU1 | 6674 | 0.219 | 0.4247 | No |
| 1182 | POLI | POLI | 6677 | 0.218 | 0.4247 | No |
| 1183 | DAZAP2 | DAZAP2 | 6678 | 0.218 | 0.4248 | No |
| 1184 | TUBGCP4 | TUBGCP4 | 6681 | 0.218 | 0.4249 | No |
| 1185 | FUZ | FUZ | 6690 | 0.217 | 0.4246 | No |
| 1186 | KMT2E | KMT2E | 6691 | 0.217 | 0.4247 | No |
| 1187 | FAM135A | FAM135A | 6696 | 0.217 | 0.4247 | No |
| 1188 | NAA16 | NAA16 | 6700 | 0.217 | 0.4246 | No |
| 1189 | MSL1 | MSL1 | 6701 | 0.217 | 0.4248 | No |
| 1190 | RNF38 | RNF38 | 6703 | 0.217 | 0.4249 | No |
| 1191 | CPEB4 | CPEB4 | 6707 | 0.216 | 0.4248 | No |
| 1192 | ABI2 | ABI2 | 6711 | 0.216 | 0.4248 | No |
| 1193 | PAIP2B | PAIP2B | 6712 | 0.216 | 0.4249 | No |
| 1194 | UBAC1 | UBAC1 | 6723 | 0.215 | 0.4246 | No |
| 1195 | SWT1 | SWT1 | 6725 | 0.215 | 0.4247 | No |
| 1196 | POU3F3 | POU3F3 | 6726 | 0.214 | 0.4248 | No |
| 1197 | TSPAN11 | TSPAN11 | 6731 | 0.213 | 0.4247 | No |
| 1198 | CNNM3 | CNNM3 | 6740 | 0.212 | 0.4244 | No |
| 1199 | FAM193A | FAM193A | 6743 | 0.211 | 0.4245 | No |
| 1200 | ABHD10 | ABHD10 | 6764 | 0.210 | 0.4236 | No |
| 1201 | LUC7L | LUC7L | 6766 | 0.210 | 0.4237 | No |
| 1202 | ZCWPW1 | ZCWPW1 | 6781 | 0.208 | 0.4231 | No |
| 1203 | CACFD1 | CACFD1 | 6797 | 0.206 | 0.4224 | No |
| 1204 | DCUN1D2 | DCUN1D2 | 6799 | 0.206 | 0.4225 | No |
| 1205 | ARRB2 | ARRB2 | 6805 | 0.205 | 0.4224 | No |
| 1206 | ANO8 | ANO8 | 6806 | 0.205 | 0.4225 | No |
| 1207 | NME4 | NME4 | 6810 | 0.205 | 0.4225 | No |
| 1208 | MROH8 | MROH8 | 6835 | 0.203 | 0.4214 | No |
| 1209 | EFCAB2 | EFCAB2 | 6847 | 0.203 | 0.4210 | No |
| 1210 | TTC33 | TTC33 | 6856 | 0.202 | 0.4207 | No |
| 1211 | ABHD17B | ABHD17B | 6857 | 0.202 | 0.4208 | No |
| 1212 | DPY19L3 | DPY19L3 | 6862 | 0.201 | 0.4207 | No |
| 1213 | MAST3 | MAST3 | 6865 | 0.201 | 0.4207 | No |
| 1214 | FANCG | FANCG | 6877 | 0.200 | 0.4203 | No |
| 1215 | TXNDC12 | TXNDC12 | 6878 | 0.200 | 0.4204 | No |
| 1216 | VPS45 | VPS45 | 6884 | 0.199 | 0.4203 | No |
| 1217 | ARHGAP42 | ARHGAP42 | 6888 | 0.199 | 0.4203 | No |
| 1218 | TRAK1 | TRAK1 | 6895 | 0.198 | 0.4201 | No |
| 1219 | SPIRE1 | SPIRE1 | 6898 | 0.198 | 0.4201 | No |
| 1220 | ABCA2 | ABCA2 | 6899 | 0.198 | 0.4202 | No |
| 1221 | CBX5 | CBX5 | 6907 | 0.197 | 0.4200 | No |
| 1222 | AGBL3 | AGBL3 | 6930 | 0.195 | 0.4190 | No |
| 1223 | FSD1 | FSD1 | 6931 | 0.195 | 0.4191 | No |
| 1224 | INPP4A | INPP4A | 6938 | 0.194 | 0.4189 | No |
| 1225 | GPT2 | GPT2 | 6949 | 0.194 | 0.4185 | No |
| 1226 | FIZ1 | FIZ1 | 6950 | 0.194 | 0.4187 | No |
| 1227 | GALNT11 | GALNT11 | 6951 | 0.194 | 0.4188 | No |
| 1228 | MYPOP | MYPOP | 6952 | 0.194 | 0.4189 | No |
| 1229 | TEX14 | TEX14 | 6956 | 0.193 | 0.4189 | No |
| 1230 | PHF2 | PHF2 | 6978 | 0.193 | 0.4179 | No |
| 1231 | TRAPPC9 | TRAPPC9 | 6984 | 0.192 | 0.4178 | No |
| 1232 | HIP1R | HIP1R | 6994 | 0.191 | 0.4174 | No |
| 1233 | HABP4 | HABP4 | 6996 | 0.191 | 0.4175 | No |
| 1234 | TRIM37 | TRIM37 | 7008 | 0.190 | 0.4171 | No |
| 1235 | BTBD2 | BTBD2 | 7019 | 0.189 | 0.4167 | No |
| 1236 | POLM | POLM | 7042 | 0.186 | 0.4157 | No |
| 1237 | PNMAL1 | PNMAL1 | 7045 | 0.186 | 0.4157 | No |
| 1238 | CBARP | CBARP | 7051 | 0.185 | 0.4155 | No |
| 1239 | FAM222B | FAM222B | 7059 | 0.184 | 0.4153 | No |
| 1240 | ITM2C | ITM2C | 7060 | 0.184 | 0.4154 | No |
| 1241 | LTK | LTK | 7066 | 0.183 | 0.4153 | No |
| 1242 | CDC42SE1 | CDC42SE1 | 7071 | 0.183 | 0.4152 | No |
| 1243 | SOGA1 | SOGA1 | 7076 | 0.182 | 0.4151 | No |
| 1244 | POU2F1 | POU2F1 | 7077 | 0.182 | 0.4152 | No |
| 1245 | ZMAT1 | ZMAT1 | 7084 | 0.182 | 0.4150 | No |
| 1246 | MTSS1L | MTSS1L | 7089 | 0.181 | 0.4149 | No |
| 1247 | PCGF3 | PCGF3 | 7123 | 0.179 | 0.4134 | No |
| 1248 | CDC25A | CDC25A | 7124 | 0.179 | 0.4135 | No |
| 1249 | FOXO3 | FOXO3 | 7126 | 0.179 | 0.4135 | No |
| 1250 | DRAM2 | DRAM2 | 7127 | 0.179 | 0.4136 | No |
| 1251 | STARD9 | STARD9 | 7129 | 0.179 | 0.4137 | No |
| 1252 | CARF | CARF | 7134 | 0.178 | 0.4136 | No |
| 1253 | KCTD3 | KCTD3 | 7140 | 0.178 | 0.4135 | No |
| 1254 | SEMA6A | SEMA6A | 7152 | 0.177 | 0.4130 | No |
| 1255 | MACROD1 | MACROD1 | 7158 | 0.177 | 0.4129 | No |
| 1256 | ITGA2B | ITGA2B | 7163 | 0.176 | 0.4128 | No |
| 1257 | SFSWAP | SFSWAP | 7164 | 0.176 | 0.4129 | No |
| 1258 | H2AFY | H2AFY | 7173 | 0.176 | 0.4126 | No |
| 1259 | FSD1L | FSD1L | 7181 | 0.175 | 0.4123 | No |
| 1260 | ZFR2 | ZFR2 | 7184 | 0.174 | 0.4123 | No |
| 1261 | CAB39L | CAB39L | 7186 | 0.174 | 0.4124 | No |
| 1262 | DACT3 | DACT3 | 7188 | 0.174 | 0.4124 | No |
| 1263 | FAM76B | FAM76B | 7189 | 0.174 | 0.4125 | No |
| 1264 | MRPS25 | MRPS25 | 7208 | 0.172 | 0.4117 | No |
| 1265 | TMEM106B | TMEM106B | 7216 | 0.172 | 0.4115 | No |
| 1266 | SLC27A3 | SLC27A3 | 7222 | 0.171 | 0.4113 | No |
| 1267 | CCNT2 | CCNT2 | 7227 | 0.171 | 0.4112 | No |
| 1268 | CNOT8 | CNOT8 | 7230 | 0.170 | 0.4112 | No |
| 1269 | HADH | HADH | 7261 | 0.167 | 0.4098 | No |
| 1270 | BRAT1 | BRAT1 | 7264 | 0.166 | 0.4098 | No |
| 1271 | SLC37A4 | SLC37A4 | 7273 | 0.166 | 0.4095 | No |
| 1272 | ARHGAP35 | ARHGAP35 | 7274 | 0.166 | 0.4096 | No |
| 1273 | MRPS26 | MRPS26 | 7278 | 0.166 | 0.4096 | No |
| 1274 | SETD5 | SETD5 | 7284 | 0.165 | 0.4094 | No |
| 1275 | SLC23A2 | SLC23A2 | 7288 | 0.164 | 0.4094 | No |
| 1276 | PCF11 | PCF11 | 7300 | 0.164 | 0.4089 | No |
| 1277 | LRRC20 | LRRC20 | 7301 | 0.164 | 0.4090 | No |
| 1278 | VGF | VGF | 7304 | 0.163 | 0.4090 | No |
| 1279 | FAM192A | FAM192A | 7306 | 0.163 | 0.4091 | No |
| 1280 | FBXO45 | FBXO45 | 7319 | 0.162 | 0.4086 | No |
| 1281 | KLHL12 | KLHL12 | 7320 | 0.162 | 0.4087 | No |
| 1282 | FAM173A | FAM173A | 7321 | 0.162 | 0.4088 | No |
| 1283 | CASC3 | CASC3 | 7328 | 0.161 | 0.4085 | No |
| 1284 | EID2 | EID2 | 7330 | 0.161 | 0.4086 | No |
| 1285 | WASF3 | WASF3 | 7346 | 0.160 | 0.4079 | No |
| 1286 | INPPL1 | INPPL1 | 7352 | 0.159 | 0.4078 | No |
| 1287 | SNRK | SNRK | 7357 | 0.159 | 0.4077 | No |
| 1288 | ALDH5A1 | ALDH5A1 | 7362 | 0.159 | 0.4076 | No |
| 1289 | SECISBP2L | SECISBP2L | 7376 | 0.158 | 0.4070 | No |
| 1290 | R3HDM4 | R3HDM4 | 7381 | 0.157 | 0.4069 | No |
| 1291 | CCDC88A | CCDC88A | 7390 | 0.156 | 0.4066 | No |
| 1292 | HSF2 | HSF2 | 7394 | 0.156 | 0.4065 | No |
| 1293 | GTF3C2 | GTF3C2 | 7404 | 0.155 | 0.4062 | No |
| 1294 | TYRO3 | TYRO3 | 7405 | 0.155 | 0.4063 | No |
| 1295 | UBL3 | UBL3 | 7410 | 0.155 | 0.4062 | No |
| 1296 | TSPAN3 | TSPAN3 | 7416 | 0.154 | 0.4060 | No |
| 1297 | CCDC40 | CCDC40 | 7418 | 0.154 | 0.4060 | No |
| 1298 | SRPK2 | SRPK2 | 7425 | 0.154 | 0.4058 | No |
| 1299 | ABCA3 | ABCA3 | 7439 | 0.153 | 0.4053 | No |
| 1300 | SLC5A6 | SLC5A6 | 7441 | 0.152 | 0.4053 | No |
| 1301 | GATAD2B | GATAD2B | 7444 | 0.152 | 0.4053 | No |
| 1302 | ZMAT3 | ZMAT3 | 7448 | 0.152 | 0.4052 | No |
| 1303 | FASTKD1 | FASTKD1 | 7452 | 0.152 | 0.4052 | No |
| 1304 | EPC1 | EPC1 | 7467 | 0.150 | 0.4046 | No |
| 1305 | CCDC159 | CCDC159 | 7481 | 0.149 | 0.4040 | No |
| 1306 | FREM2 | FREM2 | 7482 | 0.149 | 0.4041 | No |
| 1307 | SNX27 | SNX27 | 7490 | 0.149 | 0.4038 | No |
| 1308 | SEPT4 | SEPT4 | 7502 | 0.148 | 0.4034 | No |
| 1309 | PDCD6 | PDCD6 | 7504 | 0.147 | 0.4034 | No |
| 1310 | CRY1 | CRY1 | 7508 | 0.147 | 0.4033 | No |
| 1311 | LRRC4 | LRRC4 | 7517 | 0.146 | 0.4030 | No |
| 1312 | INPP5B | INPP5B | 7528 | 0.145 | 0.4026 | No |
| 1313 | CHCHD6 | CHCHD6 | 7533 | 0.145 | 0.4025 | No |
| 1314 | STAT5B | STAT5B | 7535 | 0.145 | 0.4025 | No |
| 1315 | TCF4 | TCF4 | 7537 | 0.144 | 0.4026 | No |
| 1316 | CBLL1 | CBLL1 | 7539 | 0.144 | 0.4026 | No |
| 1317 | WRAP73 | WRAP73 | 7543 | 0.144 | 0.4025 | No |
| 1318 | DHX30 | DHX30 | 7550 | 0.144 | 0.4023 | No |
| 1319 | MAPK10 | MAPK10 | 7553 | 0.143 | 0.4023 | No |
| 1320 | SSBP4 | SSBP4 | 7556 | 0.143 | 0.4023 | No |
| 1321 | KMT2B | KMT2B | 7562 | 0.143 | 0.4021 | No |
| 1322 | ATP2A1 | ATP2A1 | 7580 | 0.142 | 0.4013 | No |
| 1323 | PARP1 | PARP1 | 7606 | 0.142 | 0.4002 | No |
| 1324 | TIGD3 | TIGD3 | 7626 | 0.141 | 0.3993 | No |
| 1325 | FAM228A | FAM228A | 7635 | 0.141 | 0.3990 | No |
| 1326 | USP21 | USP21 | 7652 | 0.141 | 0.3982 | No |
| 1327 | DDR1 | DDR1 | 7658 | 0.140 | 0.3981 | No |
| 1328 | DHODH | DHODH | 7660 | 0.140 | 0.3981 | No |
| 1329 | RTN3 | RTN3 | 7662 | 0.140 | 0.3981 | No |
| 1330 | TMEM147 | TMEM147 | 7663 | 0.140 | 0.3982 | No |
| 1331 | STARD7 | STARD7 | 7668 | 0.139 | 0.3981 | No |
| 1332 | CALM3 | CALM3 | 7669 | 0.139 | 0.3982 | No |
| 1333 | WDR17 | WDR17 | 7672 | 0.139 | 0.3982 | No |
| 1334 | SLC25A36 | SLC25A36 | 7675 | 0.139 | 0.3982 | No |
| 1335 | TARS2 | TARS2 | 7686 | 0.138 | 0.3977 | No |
| 1336 | TMEM121 | TMEM121 | 7693 | 0.138 | 0.3975 | No |
| 1337 | HUNK | HUNK | 7697 | 0.138 | 0.3975 | No |
| 1338 | GPR137B | GPR137B | 7707 | 0.137 | 0.3971 | No |
| 1339 | WDR60 | WDR60 | 7710 | 0.137 | 0.3971 | No |
| 1340 | PPM1L | PPM1L | 7716 | 0.137 | 0.3969 | No |
| 1341 | LAT2 | LAT2 | 7722 | 0.136 | 0.3967 | No |
| 1342 | RAB3B | RAB3B | 7727 | 0.136 | 0.3966 | No |
| 1343 | RPRD2 | RPRD2 | 7731 | 0.135 | 0.3965 | No |
| 1344 | IFT81 | IFT81 | 7735 | 0.135 | 0.3965 | No |
| 1345 | TMEM201 | TMEM201 | 7738 | 0.135 | 0.3965 | No |
| 1346 | SDHAF1 | SDHAF1 | 7744 | 0.135 | 0.3963 | No |
| 1347 | CCDC61 | CCDC61 | 7745 | 0.134 | 0.3964 | No |
| 1348 | SMARCD3 | SMARCD3 | 7748 | 0.134 | 0.3963 | No |
| 1349 | VAV3 | VAV3 | 7750 | 0.134 | 0.3964 | No |
| 1350 | MAMLD1 | MAMLD1 | 7753 | 0.134 | 0.3964 | No |
| 1351 | ACO2 | ACO2 | 7758 | 0.133 | 0.3962 | No |
| 1352 | TMEM170B | TMEM170B | 7760 | 0.133 | 0.3963 | No |
| 1353 | TET3 | TET3 | 7768 | 0.133 | 0.3960 | No |
| 1354 | DEF8 | DEF8 | 7772 | 0.132 | 0.3959 | No |
| 1355 | SNRPA | SNRPA | 7813 | 0.131 | 0.3940 | No |
| 1356 | STRN4 | STRN4 | 7836 | 0.130 | 0.3929 | No |
| 1357 | CHTOP | CHTOP | 7840 | 0.130 | 0.3929 | No |
| 1358 | RMND5A | RMND5A | 7849 | 0.130 | 0.3925 | No |
| 1359 | MTX3 | MTX3 | 7860 | 0.129 | 0.3921 | No |
| 1360 | SUCLG1 | SUCLG1 | 7865 | 0.128 | 0.3920 | No |
| 1361 | HDHD2 | HDHD2 | 7868 | 0.128 | 0.3920 | No |
| 1362 | GNB4 | GNB4 | 7871 | 0.128 | 0.3919 | No |
| 1363 | DLG4 | DLG4 | 7883 | 0.127 | 0.3915 | No |
| 1364 | ARL8A | ARL8A | 7885 | 0.127 | 0.3915 | No |
| 1365 | EXOC4 | EXOC4 | 7886 | 0.127 | 0.3916 | No |
| 1366 | BRMS1L | BRMS1L | 7895 | 0.126 | 0.3912 | No |
| 1367 | RAB6B | RAB6B | 7897 | 0.126 | 0.3913 | No |
| 1368 | HDAC5 | HDAC5 | 7898 | 0.126 | 0.3913 | No |
| 1369 | PDIK1L | PDIK1L | 7901 | 0.126 | 0.3913 | No |
| 1370 | DZIP3 | DZIP3 | 7907 | 0.125 | 0.3911 | No |
| 1371 | AHSA2 | AHSA2 | 7908 | 0.125 | 0.3912 | No |
| 1372 | PAFAH1B3 | PAFAH1B3 | 7917 | 0.125 | 0.3909 | No |
| 1373 | NGDN | NGDN | 7921 | 0.125 | 0.3908 | No |
| 1374 | GPATCH11 | GPATCH11 | 7922 | 0.124 | 0.3909 | No |
| 1375 | HEXIM2 | HEXIM2 | 7924 | 0.124 | 0.3909 | No |
| 1376 | SCAPER | SCAPER | 7926 | 0.124 | 0.3909 | No |
| 1377 | TRIM24 | TRIM24 | 7942 | 0.123 | 0.3903 | No |
| 1378 | ENDOG | ENDOG | 7945 | 0.123 | 0.3902 | No |
| 1379 | PDE6D | PDE6D | 7954 | 0.123 | 0.3899 | No |
| 1380 | MAPK8IP3 | MAPK8IP3 | 7965 | 0.122 | 0.3895 | No |
| 1381 | THAP2 | THAP2 | 7979 | 0.121 | 0.3889 | No |
| 1382 | MIOS | MIOS | 7991 | 0.120 | 0.3884 | No |
| 1383 | LSM11 | LSM11 | 7992 | 0.120 | 0.3885 | No |
| 1384 | ZFYVE9 | ZFYVE9 | 8007 | 0.119 | 0.3878 | No |
| 1385 | PON2 | PON2 | 8017 | 0.118 | 0.3875 | No |
| 1386 | HNRNPA0 | HNRNPA0 | 8019 | 0.118 | 0.3875 | No |
| 1387 | PUM2 | PUM2 | 8039 | 0.117 | 0.3866 | No |
| 1388 | NKTR | NKTR | 8052 | 0.116 | 0.3860 | No |
| 1389 | SNRNP70 | SNRNP70 | 8055 | 0.116 | 0.3860 | No |
| 1390 | KDM4A | KDM4A | 8059 | 0.115 | 0.3859 | No |
| 1391 | COQ9 | COQ9 | 8069 | 0.115 | 0.3855 | No |
| 1392 | KCTD7 | KCTD7 | 8080 | 0.114 | 0.3851 | No |
| 1393 | STRADA | STRADA | 8091 | 0.113 | 0.3847 | No |
| 1394 | SMARCA4 | SMARCA4 | 8100 | 0.112 | 0.3843 | No |
| 1395 | MLF1 | MLF1 | 8106 | 0.111 | 0.3842 | No |
| 1396 | CSTF2T | CSTF2T | 8118 | 0.111 | 0.3837 | No |
| 1397 | CBX4 | CBX4 | 8126 | 0.110 | 0.3834 | No |
| 1398 | NUP133 | NUP133 | 8130 | 0.109 | 0.3833 | No |
| 1399 | NUDT1 | NUDT1 | 8131 | 0.109 | 0.3834 | No |
| 1400 | SUGP2 | SUGP2 | 8133 | 0.109 | 0.3834 | No |
| 1401 | SNX4 | SNX4 | 8151 | 0.108 | 0.3826 | No |
| 1402 | NLK | NLK | 8158 | 0.107 | 0.3823 | No |
| 1403 | RNF216 | RNF216 | 8165 | 0.107 | 0.3821 | No |
| 1404 | RSBN1L | RSBN1L | 8173 | 0.107 | 0.3818 | No |
| 1405 | AGAP1 | AGAP1 | 8180 | 0.106 | 0.3816 | No |
| 1406 | WRB | WRB | 8184 | 0.106 | 0.3815 | No |
| 1407 | ECI1 | ECI1 | 8192 | 0.105 | 0.3812 | No |
| 1408 | LMBR1 | LMBR1 | 8220 | 0.104 | 0.3799 | No |
| 1409 | TADA2A | TADA2A | 8223 | 0.104 | 0.3799 | No |
| 1410 | DGAT1 | DGAT1 | 8227 | 0.104 | 0.3798 | No |
| 1411 | CPE | CPE | 8228 | 0.104 | 0.3798 | No |
| 1412 | PAIP2 | PAIP2 | 8238 | 0.103 | 0.3794 | No |
| 1413 | MIF4GD | MIF4GD | 8241 | 0.103 | 0.3794 | No |
| 1414 | PPP1R42 | PPP1R42 | 8242 | 0.103 | 0.3795 | No |
| 1415 | MIDN | MIDN | 8246 | 0.103 | 0.3794 | No |
| 1416 | SCARB2 | SCARB2 | 8247 | 0.103 | 0.3794 | No |
| 1417 | CACNG7 | CACNG7 | 8249 | 0.102 | 0.3794 | No |
| 1418 | MLST8 | MLST8 | 8261 | 0.101 | 0.3789 | No |
| 1419 | MATR3 | MATR3 | 8281 | 0.100 | 0.3780 | No |
| 1420 | CSPG4 | CSPG4 | 8286 | 0.099 | 0.3779 | No |
| 1421 | IDH1 | IDH1 | 8292 | 0.099 | 0.3777 | No |
| 1422 | PID1 | PID1 | 8312 | 0.098 | 0.3768 | No |
| 1423 | MEX3A | MEX3A | 8313 | 0.098 | 0.3769 | No |
| 1424 | DNAJC18 | DNAJC18 | 8319 | 0.097 | 0.3767 | No |
| 1425 | MPND | MPND | 8331 | 0.097 | 0.3762 | No |
| 1426 | ENO3 | ENO3 | 8339 | 0.096 | 0.3759 | No |
| 1427 | WNK3 | WNK3 | 8347 | 0.096 | 0.3756 | No |
| 1428 | FARP1 | FARP1 | 8349 | 0.095 | 0.3756 | No |
| 1429 | KHSRP | KHSRP | 8360 | 0.095 | 0.3751 | No |
| 1430 | DCAF7 | DCAF7 | 8369 | 0.094 | 0.3748 | No |
| 1431 | DYRK1A | DYRK1A | 8371 | 0.094 | 0.3748 | No |
| 1432 | SRI | SRI | 8372 | 0.094 | 0.3749 | No |
| 1433 | KIF3A | KIF3A | 8374 | 0.094 | 0.3749 | No |
| 1434 | SMDT1 | SMDT1 | 8375 | 0.094 | 0.3749 | No |
| 1435 | RAC1 | RAC1 | 8376 | 0.094 | 0.3750 | No |
| 1436 | ANGEL2 | ANGEL2 | 8386 | 0.093 | 0.3746 | No |
| 1437 | FOXN2 | FOXN2 | 8396 | 0.093 | 0.3742 | No |
| 1438 | STRIP1 | STRIP1 | 8398 | 0.093 | 0.3742 | No |
| 1439 | ILF3 | ILF3 | 8403 | 0.092 | 0.3740 | No |
| 1440 | REEP2 | REEP2 | 8405 | 0.092 | 0.3741 | No |
| 1441 | SCAP | SCAP | 8409 | 0.092 | 0.3740 | No |
| 1442 | CHL1 | CHL1 | 8411 | 0.092 | 0.3740 | No |
| 1443 | COA5 | COA5 | 8417 | 0.091 | 0.3738 | No |
| 1444 | PRR12 | PRR12 | 8421 | 0.091 | 0.3737 | No |
| 1445 | SLC25A44 | SLC25A44 | 8432 | 0.090 | 0.3732 | No |
| 1446 | SP6 | SP6 | 8438 | 0.090 | 0.3730 | No |
| 1447 | LRP8 | LRP8 | 8455 | 0.089 | 0.3723 | No |
| 1448 | KNOP1 | KNOP1 | 8457 | 0.089 | 0.3723 | No |
| 1449 | LIX1L | LIX1L | 8462 | 0.089 | 0.3721 | No |
| 1450 | CHD4 | CHD4 | 8463 | 0.089 | 0.3722 | No |
| 1451 | PIWIL2 | PIWIL2 | 8476 | 0.088 | 0.3716 | No |
| 1452 | SLC25A4 | SLC25A4 | 8483 | 0.088 | 0.3714 | No |
| 1453 | CORO7 | CORO7 | 8490 | 0.087 | 0.3711 | No |
| 1454 | SNX10 | SNX10 | 8494 | 0.087 | 0.3710 | No |
| 1455 | HOOK2 | HOOK2 | 8499 | 0.087 | 0.3709 | No |
| 1456 | PLEKHA8 | PLEKHA8 | 8502 | 0.087 | 0.3708 | No |
| 1457 | TMBIM6 | TMBIM6 | 8503 | 0.087 | 0.3709 | No |
| 1458 | MTMR3 | MTMR3 | 8508 | 0.086 | 0.3707 | No |
| 1459 | PRKAR1B | PRKAR1B | 8512 | 0.086 | 0.3706 | No |
| 1460 | POMGNT2 | POMGNT2 | 8521 | 0.085 | 0.3703 | No |
| 1461 | TOMM20 | TOMM20 | 8541 | 0.084 | 0.3694 | No |
| 1462 | ZBTB14 | ZBTB14 | 8550 | 0.083 | 0.3690 | No |
| 1463 | FAM53C | FAM53C | 8578 | 0.081 | 0.3677 | No |
| 1464 | KLHL22 | KLHL22 | 8579 | 0.081 | 0.3677 | No |
| 1465 | DHRS13 | DHRS13 | 8581 | 0.081 | 0.3677 | No |
| 1466 | GIGYF1 | GIGYF1 | 8593 | 0.080 | 0.3672 | No |
| 1467 | ZC3H6 | ZC3H6 | 8594 | 0.080 | 0.3673 | No |
| 1468 | MARCH6 | MARCH6 | 8595 | 0.080 | 0.3673 | No |
| 1469 | TNPO2 | TNPO2 | 8597 | 0.080 | 0.3673 | No |
| 1470 | PREX1 | PREX1 | 8599 | 0.080 | 0.3673 | No |
| 1471 | POLR1B | POLR1B | 8602 | 0.080 | 0.3673 | No |
| 1472 | ZDHHC17 | ZDHHC17 | 8630 | 0.077 | 0.3660 | No |
| 1473 | SMAP1 | SMAP1 | 8634 | 0.077 | 0.3658 | No |
| 1474 | FAM98C | FAM98C | 8638 | 0.077 | 0.3657 | No |
| 1475 | ZFP1 | ZFP1 | 8645 | 0.076 | 0.3655 | No |
| 1476 | PROCA1 | PROCA1 | 8665 | 0.075 | 0.3646 | No |
| 1477 | NT5C3B | NT5C3B | 8678 | 0.075 | 0.3640 | No |
| 1478 | GPBP1 | GPBP1 | 8693 | 0.074 | 0.3633 | No |
| 1479 | NR4A1 | NR4A1 | 8730 | 0.071 | 0.3616 | No |
| 1480 | FNDC5 | FNDC5 | 8739 | 0.071 | 0.3612 | No |
| 1481 | PMS2 | PMS2 | 8743 | 0.071 | 0.3611 | No |
| 1482 | GPSM2 | GPSM2 | 8746 | 0.071 | 0.3610 | No |
| 1483 | PTMS | PTMS | 8747 | 0.071 | 0.3611 | No |
| 1484 | ZC3H4 | ZC3H4 | 8755 | 0.070 | 0.3608 | No |
| 1485 | DUSP22 | DUSP22 | 8763 | 0.069 | 0.3605 | No |
| 1486 | PIK3R2 | PIK3R2 | 8764 | 0.069 | 0.3605 | No |
| 1487 | FBXW7 | FBXW7 | 8774 | 0.068 | 0.3601 | No |
| 1488 | PSTK | PSTK | 8779 | 0.068 | 0.3599 | No |
| 1489 | DCAF15 | DCAF15 | 8781 | 0.068 | 0.3599 | No |
| 1490 | B4GAT1 | B4GAT1 | 8799 | 0.066 | 0.3591 | No |
| 1491 | WDR35 | WDR35 | 8807 | 0.066 | 0.3588 | No |
| 1492 | PPP1R32 | PPP1R32 | 8827 | 0.065 | 0.3579 | No |
| 1493 | RSBN1 | RSBN1 | 8846 | 0.064 | 0.3570 | No |
| 1494 | POGZ | POGZ | 8865 | 0.062 | 0.3561 | No |
| 1495 | TTLL4 | TTLL4 | 8873 | 0.061 | 0.3558 | No |
| 1496 | STX6 | STX6 | 8875 | 0.061 | 0.3558 | No |
| 1497 | DUSP16 | DUSP16 | 8883 | 0.061 | 0.3555 | No |
| 1498 | EXTL3 | EXTL3 | 8890 | 0.060 | 0.3552 | No |
| 1499 | HNRNPLL | HNRNPLL | 8897 | 0.060 | 0.3549 | No |
| 1500 | HMGXB4 | HMGXB4 | 8901 | 0.060 | 0.3548 | No |
| 1501 | COQ7 | COQ7 | 8922 | 0.058 | 0.3538 | No |
| 1502 | FAM221A | FAM221A | 8935 | 0.057 | 0.3533 | No |
| 1503 | PRR14 | PRR14 | 8939 | 0.057 | 0.3532 | No |
| 1504 | RAD52 | RAD52 | 8948 | 0.056 | 0.3528 | No |
| 1505 | UBALD2 | UBALD2 | 8955 | 0.056 | 0.3525 | No |
| 1506 | FBXW11 | FBXW11 | 8958 | 0.056 | 0.3524 | No |
| 1507 | BMPR1A | BMPR1A | 8960 | 0.055 | 0.3524 | No |
| 1508 | SARAF | SARAF | 8962 | 0.055 | 0.3524 | No |
| 1509 | NDUFS3 | NDUFS3 | 8965 | 0.055 | 0.3523 | No |
| 1510 | PRDM2 | PRDM2 | 8968 | 0.055 | 0.3523 | No |
| 1511 | MSL2 | MSL2 | 8970 | 0.055 | 0.3523 | No |
| 1512 | RAB11FIP3 | RAB11FIP3 | 8975 | 0.054 | 0.3521 | No |
| 1513 | FRS2 | FRS2 | 9006 | 0.052 | 0.3506 | No |
| 1514 | MEAF6 | MEAF6 | 9024 | 0.051 | 0.3498 | No |
| 1515 | RHOT1 | RHOT1 | 9032 | 0.050 | 0.3494 | No |
| 1516 | ACAD8 | ACAD8 | 9033 | 0.050 | 0.3495 | No |
| 1517 | ACKR3 | ACKR3 | 9040 | 0.050 | 0.3492 | No |
| 1518 | COX6B1 | COX6B1 | 9048 | 0.049 | 0.3489 | No |
| 1519 | ZEB1 | ZEB1 | 9049 | 0.049 | 0.3489 | No |
| 1520 | RPS11 | RPS11 | 9050 | 0.049 | 0.3489 | No |
| 1521 | FAM171B | FAM171B | 9052 | 0.049 | 0.3489 | No |
| 1522 | ABHD17C | ABHD17C | 9053 | 0.049 | 0.3490 | No |
| 1523 | RNF44 | RNF44 | 9061 | 0.049 | 0.3486 | No |
| 1524 | RLF | RLF | 9063 | 0.049 | 0.3486 | No |
| 1525 | RAB29 | RAB29 | 9074 | 0.048 | 0.3481 | No |
| 1526 | LINGO3 | LINGO3 | 9078 | 0.047 | 0.3480 | No |
| 1527 | KANK1 | KANK1 | 9092 | 0.047 | 0.3474 | No |
| 1528 | FANCE | FANCE | 9093 | 0.047 | 0.3474 | No |
| 1529 | CABIN1 | CABIN1 | 9102 | 0.046 | 0.3470 | No |
| 1530 | MED9 | MED9 | 9107 | 0.046 | 0.3469 | No |
| 1531 | CEP68 | CEP68 | 9118 | 0.045 | 0.3464 | No |
| 1532 | ACBD6 | ACBD6 | 9119 | 0.045 | 0.3464 | No |
| 1533 | ACYP2 | ACYP2 | 9125 | 0.044 | 0.3462 | No |
| 1534 | UBXN2A | UBXN2A | 9128 | 0.044 | 0.3461 | No |
| 1535 | BRD8 | BRD8 | 9138 | 0.044 | 0.3457 | No |
| 1536 | LMO4 | LMO4 | 9156 | 0.043 | 0.3448 | No |
| 1537 | GFOD2 | GFOD2 | 9157 | 0.043 | 0.3449 | No |
| 1538 | KDM6B | KDM6B | 9158 | 0.043 | 0.3449 | No |
| 1539 | ADRB1 | ADRB1 | 9159 | 0.043 | 0.3449 | No |
| 1540 | SPPL3 | SPPL3 | 9174 | 0.041 | 0.3442 | No |
| 1541 | ATP5S | ATP5S | 9177 | 0.041 | 0.3442 | No |
| 1542 | STK16 | STK16 | 9178 | 0.041 | 0.3442 | No |
| 1543 | RPL18A | RPL18A | 9179 | 0.041 | 0.3442 | No |
| 1544 | COG2 | COG2 | 9181 | 0.041 | 0.3442 | No |
| 1545 | RSRC1 | RSRC1 | 9190 | 0.041 | 0.3438 | No |
| 1546 | SMIM8 | SMIM8 | 9203 | 0.040 | 0.3432 | No |
| 1547 | KBTBD2 | KBTBD2 | 9214 | 0.039 | 0.3427 | No |
| 1548 | FAM110B | FAM110B | 9219 | 0.038 | 0.3426 | No |
| 1549 | PHF7 | PHF7 | 9255 | 0.036 | 0.3408 | No |
| 1550 | BRWD1 | BRWD1 | 9258 | 0.036 | 0.3407 | No |
| 1551 | VPS72 | VPS72 | 9260 | 0.036 | 0.3407 | No |
| 1552 | POLR2I | POLR2I | 9273 | 0.035 | 0.3401 | No |
| 1553 | RNF187 | RNF187 | 9278 | 0.035 | 0.3399 | No |
| 1554 | CLPP | CLPP | 9310 | 0.031 | 0.3384 | No |
| 1555 | TTC32 | TTC32 | 9320 | 0.031 | 0.3379 | No |
| 1556 | UBXN8 | UBXN8 | 9344 | 0.029 | 0.3368 | No |
| 1557 | DCP2 | DCP2 | 9345 | 0.029 | 0.3368 | No |
| 1558 | NDUFS7 | NDUFS7 | 9351 | 0.029 | 0.3366 | No |
| 1559 | SMARCC2 | SMARCC2 | 9366 | 0.028 | 0.3359 | No |
| 1560 | STMN4 | STMN4 | 9371 | 0.027 | 0.3357 | No |
| 1561 | TFAP4 | TFAP4 | 9372 | 0.027 | 0.3357 | No |
| 1562 | BCDIN3D | BCDIN3D | 9378 | 0.027 | 0.3355 | No |
| 1563 | REPIN1 | REPIN1 | 9381 | 0.027 | 0.3354 | No |
| 1564 | YEATS4 | YEATS4 | 9393 | 0.026 | 0.3349 | No |
| 1565 | PRPS1 | PRPS1 | 9395 | 0.026 | 0.3348 | No |
| 1566 | FEM1B | FEM1B | 9410 | 0.025 | 0.3341 | No |
| 1567 | TRMT10A | TRMT10A | 9415 | 0.024 | 0.3339 | No |
| 1568 | NELFA | NELFA | 9417 | 0.024 | 0.3339 | No |
| 1569 | MTA1 | MTA1 | 9431 | 0.023 | 0.3333 | No |
| 1570 | CHERP | CHERP | 9433 | 0.023 | 0.3332 | No |
| 1571 | AHCYL2 | AHCYL2 | 9437 | 0.023 | 0.3331 | No |
| 1572 | MAP3K10 | MAP3K10 | 9439 | 0.023 | 0.3331 | No |
| 1573 | PIP4K2B | PIP4K2B | 9443 | 0.023 | 0.3329 | No |
| 1574 | CALM1 | CALM1 | 9467 | 0.021 | 0.3318 | No |
| 1575 | DIP2C | DIP2C | 9470 | 0.021 | 0.3317 | No |
| 1576 | TIMM50 | TIMM50 | 9488 | 0.019 | 0.3308 | No |
| 1577 | DNER | DNER | 9498 | 0.018 | 0.3304 | No |
| 1578 | DTNA | DTNA | 9500 | 0.018 | 0.3303 | No |
| 1579 | MDM1 | MDM1 | 9512 | 0.017 | 0.3298 | No |
| 1580 | ECSIT | ECSIT | 9521 | 0.017 | 0.3294 | No |
| 1581 | KCNAB3 | KCNAB3 | 9524 | 0.017 | 0.3293 | No |
| 1582 | LZTS3 | LZTS3 | 9535 | 0.017 | 0.3288 | No |
| 1583 | HECTD2 | HECTD2 | 9537 | 0.016 | 0.3288 | No |
| 1584 | RP9 | RP9 | 9539 | 0.016 | 0.3287 | No |
| 1585 | HEPACAM2 | HEPACAM2 | 9558 | 0.015 | 0.3278 | No |
| 1586 | JMJD7 | JMJD7 | 9560 | 0.015 | 0.3278 | No |
| 1587 | MAP2K5 | MAP2K5 | 9567 | 0.014 | 0.3275 | No |
| 1588 | NDUFA4 | NDUFA4 | 9571 | 0.014 | 0.3274 | No |
| 1589 | AKAP8L | AKAP8L | 9600 | 0.012 | 0.3259 | No |
| 1590 | ATP5A1 | ATP5A1 | 9604 | 0.012 | 0.3258 | No |
| 1591 | POMT2 | POMT2 | 9612 | 0.012 | 0.3254 | No |
| 1592 | ARID4A | ARID4A | 9614 | 0.011 | 0.3254 | No |
| 1593 | IFFO1 | IFFO1 | 9619 | 0.011 | 0.3252 | No |
| 1594 | LUC7L3 | LUC7L3 | 9627 | 0.010 | 0.3249 | No |
| 1595 | OTUD7A | OTUD7A | 9641 | 0.010 | 0.3242 | No |
| 1596 | TECTA | TECTA | 9645 | 0.010 | 0.3241 | No |
| 1597 | TSSK6 | TSSK6 | 9648 | 0.010 | 0.3240 | No |
| 1598 | ADAM1A | ADAM1A | 9658 | 0.010 | 0.3235 | No |
| 1599 | LST1 | LST1 | 9660 | 0.010 | 0.3235 | No |
| 1600 | LIN7B | LIN7B | 9723 | 0.009 | 0.3203 | No |
| 1601 | TRDN | TRDN | 9730 | 0.009 | 0.3200 | No |
| 1602 | TFR2 | TFR2 | 9734 | 0.009 | 0.3199 | No |
| 1603 | PHF24 | PHF24 | 9740 | 0.009 | 0.3197 | No |
| 1604 | SYNGR4 | SYNGR4 | 9741 | 0.009 | 0.3197 | No |
| 1605 | FAM221B | FAM221B | 9744 | 0.009 | 0.3196 | No |
| 1606 | SLC9B1 | SLC9B1 | 9747 | 0.009 | 0.3195 | No |
| 1607 | BRICD5 | BRICD5 | 9818 | 0.008 | 0.3159 | No |
| 1608 | CEP192 | CEP192 | 9836 | 0.008 | 0.3151 | No |
| 1609 | AGAP3 | AGAP3 | 9861 | 0.007 | 0.3139 | No |
| 1610 | ANKRD10 | ANKRD10 | 9870 | 0.006 | 0.3135 | No |
| 1611 | BAHD1 | BAHD1 | 9872 | 0.006 | 0.3134 | No |
| 1612 | OVGP1 | OVGP1 | 9879 | 0.006 | 0.3131 | No |
| 1613 | VOPP1 | VOPP1 | 9883 | 0.005 | 0.3130 | No |
| 1614 | NBEA | NBEA | 9884 | 0.005 | 0.3130 | No |
| 1615 | GKAP1 | GKAP1 | 9886 | 0.005 | 0.3129 | No |
| 1616 | UCK2 | UCK2 | 9890 | 0.005 | 0.3128 | No |
| 1617 | CHD1 | CHD1 | 9891 | 0.005 | 0.3128 | No |
| 1618 | KIF2A | KIF2A | 9897 | 0.004 | 0.3125 | No |
| 1619 | HERPUD2 | HERPUD2 | 9907 | 0.003 | 0.3121 | No |
| 1620 | NARFL | NARFL | 9911 | 0.003 | 0.3119 | No |
| 1621 | IRAK1BP1 | IRAK1BP1 | 9912 | 0.003 | 0.3119 | No |
| 1622 | HES7 | HES7 | 9922 | 0.003 | 0.3115 | No |
| 1623 | UQCRFS1 | UQCRFS1 | 9928 | 0.003 | 0.3112 | No |
| 1624 | DNAJC27 | DNAJC27 | 9957 | 0.000 | 0.3098 | No |
| 1625 | UNC5CL | UNC5CL | 9981 | 0.000 | 0.3086 | No |
| 1626 | ADAMTS3 | ADAMTS3 | 9982 | 0.000 | 0.3086 | No |
| 1627 | VWCE | VWCE | 9988 | 0.000 | 0.3084 | No |
| 1628 | S1PR5 | S1PR5 | 10018 | 0.000 | 0.3069 | No |
| 1629 | USH1G | USH1G | 10025 | 0.000 | 0.3066 | No |
| 1630 | HPCAL4 | HPCAL4 | 10045 | 0.000 | 0.3057 | No |
| 1631 | RPRML | RPRML | 10047 | 0.000 | 0.3056 | No |
| 1632 | GPR61 | GPR61 | 10056 | 0.000 | 0.3052 | No |
| 1633 | CHST9 | CHST9 | 10063 | 0.000 | 0.3049 | No |
| 1634 | ZBTB8B | ZBTB8B | 10082 | 0.000 | 0.3040 | No |
| 1635 | AKNAD1 | AKNAD1 | 10109 | 0.000 | 0.3027 | No |
| 1636 | PRSS46 | PRSS46 | 10112 | 0.000 | 0.3026 | No |
| 1637 | ANKRD55 | ANKRD55 | 10116 | 0.000 | 0.3024 | No |
| 1638 | CBY3 | CBY3 | 10119 | 0.000 | 0.3023 | No |
| 1639 | VSTM4 | VSTM4 | 10130 | 0.000 | 0.3018 | No |
| 1640 | PRSS27 | PRSS27 | 10132 | 0.000 | 0.3018 | No |
| 1641 | SSTR5 | SSTR5 | 10133 | 0.000 | 0.3018 | No |
| 1642 | MAP3K19 | MAP3K19 | 10148 | 0.000 | 0.3010 | No |
| 1643 | SEC31B | SEC31B | 10154 | 0.000 | 0.3008 | No |
| 1644 | DOC2A | DOC2A | 10164 | 0.000 | 0.3003 | No |
| 1645 | ACTN2 | ACTN2 | 10165 | 0.000 | 0.3003 | No |
| 1646 | CPVL | CPVL | 10176 | 0.000 | 0.2998 | No |
| 1647 | GSX1 | GSX1 | 10180 | 0.000 | 0.2997 | No |
| 1648 | UMODL1 | UMODL1 | 10194 | 0.000 | 0.2990 | No |
| 1649 | SLC6A2 | SLC6A2 | 10220 | 0.000 | 0.2978 | No |
| 1650 | LMAN1L | LMAN1L | 10229 | 0.000 | 0.2974 | No |
| 1651 | RIT2 | RIT2 | 10248 | 0.000 | 0.2964 | No |
| 1652 | IL1RAPL2 | IL1RAPL2 | 10271 | 0.000 | 0.2953 | No |
| 1653 | CDH24 | CDH24 | 10282 | 0.000 | 0.2948 | No |
| 1654 | MYL3 | MYL3 | 10283 | 0.000 | 0.2948 | No |
| 1655 | KCNG2 | KCNG2 | 10284 | 0.000 | 0.2948 | No |
| 1656 | SNTB1 | SNTB1 | 10294 | 0.000 | 0.2944 | No |
| 1657 | AGBL4 | AGBL4 | 10307 | 0.000 | 0.2938 | No |
| 1658 | ODF1 | ODF1 | 10317 | 0.000 | 0.2933 | No |
| 1659 | WSCD2 | WSCD2 | 10337 | 0.000 | 0.2923 | No |
| 1660 | JAKMIP1 | JAKMIP1 | 10340 | 0.000 | 0.2922 | No |
| 1661 | IZUMO1 | IZUMO1 | 10347 | 0.000 | 0.2919 | No |
| 1662 | PLCXD1 | PLCXD1 | 10349 | 0.000 | 0.2919 | No |
| 1663 | GIMAP8 | GIMAP8 | 10350 | 0.000 | 0.2919 | No |
| 1664 | SNORD42B | SNORD42B | 10390 | 0.000 | 0.2899 | No |
| 1665 | TNFRSF13C | TNFRSF13C | 10426 | 0.000 | 0.2881 | No |
| 1666 | MYBPHL | MYBPHL | 10432 | 0.000 | 0.2879 | No |
| 1667 | CCNB1IP1 | CCNB1IP1 | 10483 | 0.000 | 0.2854 | No |
| 1668 | KLF12 | KLF12 | 10491 | 0.000 | 0.2850 | No |
| 1669 | TMEM253 | TMEM253 | 10497 | 0.000 | 0.2848 | No |
| 1670 | MYH7B | MYH7B | 10553 | 0.000 | 0.2820 | No |
| 1671 | EIF4E1B | EIF4E1B | 10562 | 0.000 | 0.2816 | No |
| 1672 | MIR92B | MIR92B | 10582 | 0.000 | 0.2806 | No |
| 1673 | ATP6AP1L | ATP6AP1L | 10621 | 0.000 | 0.2787 | No |
| 1674 | ETV2 | ETV2 | 10786 | 0.000 | 0.2704 | No |
| 1675 | ATP1A4 | ATP1A4 | 10793 | 0.000 | 0.2701 | No |
| 1676 | MC5R | MC5R | 10794 | 0.000 | 0.2701 | No |
| 1677 | TPPP2 | TPPP2 | 10797 | 0.000 | 0.2700 | No |
| 1678 | KCNQ1 | KCNQ1 | 10803 | 0.000 | 0.2697 | No |
| 1679 | PLIN5 | PLIN5 | 10810 | 0.000 | 0.2694 | No |
| 1680 | CPB1 | CPB1 | 10811 | 0.000 | 0.2694 | No |
| 1681 | SNX31 | SNX31 | 10818 | 0.000 | 0.2691 | No |
| 1682 | IL17RB | IL17RB | 10837 | 0.000 | 0.2682 | No |
| 1683 | CACNA1D | CACNA1D | 10838 | 0.000 | 0.2682 | No |
| 1684 | CHDH | CHDH | 10839 | 0.000 | 0.2682 | No |
| 1685 | EEF1A2 | EEF1A2 | 10842 | 0.000 | 0.2681 | No |
| 1686 | YBX2 | YBX2 | 10852 | 0.000 | 0.2676 | No |
| 1687 | SLC2A4 | SLC2A4 | 10853 | 0.000 | 0.2676 | No |
| 1688 | CLVS2 | CLVS2 | 10863 | 0.000 | 0.2672 | No |
| 1689 | GRM1 | GRM1 | 10864 | 0.000 | 0.2672 | No |
| 1690 | HAVCR2 | HAVCR2 | 10885 | 0.000 | 0.2662 | No |
| 1691 | CFAP52 | CFAP52 | 10901 | 0.000 | 0.2654 | No |
| 1692 | CCDC175 | CCDC175 | 10904 | 0.000 | 0.2653 | No |
| 1693 | AOAH | AOAH | 10907 | 0.000 | 0.2652 | No |
| 1694 | SCGN | SCGN | 10908 | 0.000 | 0.2652 | No |
| 1695 | TRPC7 | TRPC7 | 10914 | 0.000 | 0.2650 | No |
| 1696 | SFTPC | SFTPC | 10932 | 0.000 | 0.2641 | No |
| 1697 | SDR39U1 | SDR39U1 | 10934 | 0.000 | 0.2641 | No |
| 1698 | AHSG | AHSG | 10949 | 0.000 | 0.2633 | No |
| 1699 | ZSCAN10 | ZSCAN10 | 10966 | 0.000 | 0.2625 | No |
| 1700 | PDIA2 | PDIA2 | 10975 | 0.000 | 0.2621 | No |
| 1701 | RGS11 | RGS11 | 10976 | 0.000 | 0.2621 | No |
| 1702 | PCDH12 | PCDH12 | 10981 | 0.000 | 0.2619 | No |
| 1703 | GALR1 | GALR1 | 10984 | 0.000 | 0.2618 | No |
| 1704 | SLIT1 | SLIT1 | 11002 | 0.000 | 0.2610 | No |
| 1705 | HABP2 | HABP2 | 11005 | 0.000 | 0.2609 | No |
| 1706 | MSTN | MSTN | 11022 | 0.000 | 0.2601 | No |
| 1707 | COL19A1 | COL19A1 | 11024 | 0.000 | 0.2600 | No |
| 1708 | CDH7 | CDH7 | 11032 | 0.000 | 0.2597 | No |
| 1709 | CHIT1 | CHIT1 | 11040 | 0.000 | 0.2593 | No |
| 1710 | MYOG | MYOG | 11041 | 0.000 | 0.2593 | No |
| 1711 | LHX4 | LHX4 | 11042 | 0.000 | 0.2593 | No |
| 1712 | LAMP5 | LAMP5 | 11068 | 0.000 | 0.2580 | No |
| 1713 | RASGRP1 | RASGRP1 | 11069 | 0.000 | 0.2580 | No |
| 1714 | OTOR | OTOR | 11073 | 0.000 | 0.2579 | No |
| 1715 | SLA2 | SLA2 | 11081 | 0.000 | 0.2575 | No |
| 1716 | PTCH2 | PTCH2 | 11111 | 0.000 | 0.2561 | No |
| 1717 | SLC30A3 | SLC30A3 | 11125 | 0.000 | 0.2554 | No |
| 1718 | YIPF7 | YIPF7 | 11127 | 0.000 | 0.2554 | No |
| 1719 | GABRA4 | GABRA4 | 11129 | 0.000 | 0.2553 | No |
| 1720 | OCM | OCM | 11146 | 0.000 | 0.2545 | No |
| 1721 | AASS | AASS | 11149 | 0.000 | 0.2544 | No |
| 1722 | EPHA1 | EPHA1 | 11156 | 0.000 | 0.2541 | No |
| 1723 | PRMT8 | PRMT8 | 11176 | 0.000 | 0.2531 | No |
| 1724 | PLIN1 | PLIN1 | 11184 | 0.000 | 0.2528 | No |
| 1725 | CCKBR | CCKBR | 11199 | 0.000 | 0.2521 | No |
| 1726 | GABRQ | GABRQ | 11212 | 0.000 | 0.2515 | No |
| 1727 | ASB11 | ASB11 | 11214 | 0.000 | 0.2514 | No |
| 1728 | MYOM2 | MYOM2 | 11216 | 0.000 | 0.2514 | No |
| 1729 | TEX29 | TEX29 | 11218 | 0.000 | 0.2513 | No |
| 1730 | CDH15 | CDH15 | 11236 | 0.000 | 0.2504 | No |
| 1731 | CAPN9 | CAPN9 | 11238 | 0.000 | 0.2504 | No |
| 1732 | MST1 | MST1 | 11256 | 0.000 | 0.2495 | No |
| 1733 | RTP1 | RTP1 | 11268 | 0.000 | 0.2490 | No |
| 1734 | NEU4 | NEU4 | 11275 | 0.000 | 0.2487 | No |
| 1735 | ATP2C2 | ATP2C2 | 11277 | 0.000 | 0.2486 | No |
| 1736 | EMID1 | EMID1 | 11280 | 0.000 | 0.2485 | No |
| 1737 | ANKMY1 | ANKMY1 | 11281 | 0.000 | 0.2485 | No |
| 1738 | TMEM132C | TMEM132C | 11283 | 0.000 | 0.2485 | No |
| 1739 | HSD17B13 | HSD17B13 | 11289 | 0.000 | 0.2482 | No |
| 1740 | KRT20 | KRT20 | 11305 | 0.000 | 0.2475 | No |
| 1741 | IGFLR1 | IGFLR1 | 11326 | 0.000 | 0.2464 | No |
| 1742 | STOX1 | STOX1 | 11327 | 0.000 | 0.2464 | No |
| 1743 | KCNH3 | KCNH3 | 11341 | 0.000 | 0.2458 | No |
| 1744 | SLC9B2 | SLC9B2 | 11349 | 0.000 | 0.2454 | No |
| 1745 | CER1 | CER1 | 11355 | 0.000 | 0.2452 | No |
| 1746 | SLC19A3 | SLC19A3 | 11360 | 0.000 | 0.2450 | No |
| 1747 | HILS1 | HILS1 | 11374 | 0.000 | 0.2443 | No |
| 1748 | CDH26 | CDH26 | 11378 | 0.000 | 0.2442 | No |
| 1749 | CPLX3 | CPLX3 | 11392 | 0.000 | 0.2435 | No |
| 1750 | NINJ2 | NINJ2 | 11421 | 0.000 | 0.2421 | No |
| 1751 | PRIMA1 | PRIMA1 | 11428 | 0.000 | 0.2418 | No |
| 1752 | MYO3B | MYO3B | 11435 | 0.000 | 0.2415 | No |
| 1753 | RNF224 | RNF224 | 11904 | 0.000 | 0.2178 | No |
| 1754 | AK9 | AK9 | 11959 | 0.000 | 0.2151 | No |
| 1755 | SMIM17 | SMIM17 | 12017 | 0.000 | 0.2122 | No |
| 1756 | SMIM18 | SMIM18 | 12045 | 0.000 | 0.2108 | No |
| 1757 | MOAP1 | MOAP1 | 12101 | 0.000 | 0.2080 | No |
| 1758 | PCDHAC2 | PCDHAC2 | 12449 | 0.000 | 0.1905 | No |
| 1759 | ZIM2 | ZIM2 | 12884 | 0.000 | 0.1685 | No |
| 1760 | NDUFS2 | NDUFS2 | 13018 | -0.001 | 0.1618 | No |
| 1761 | EQTN | EQTN | 13037 | -0.001 | 0.1609 | No |
| 1762 | ROBO3 | ROBO3 | 13039 | -0.001 | 0.1608 | No |
| 1763 | PSMG3 | PSMG3 | 13104 | -0.001 | 0.1576 | No |
| 1764 | SYNPR | SYNPR | 13116 | -0.002 | 0.1570 | No |
| 1765 | ADIPOQ | ADIPOQ | 13129 | -0.002 | 0.1564 | No |
| 1766 | SNTG1 | SNTG1 | 13131 | -0.002 | 0.1564 | No |
| 1767 | FAM166A | FAM166A | 13132 | -0.002 | 0.1564 | No |
| 1768 | CNKSR1 | CNKSR1 | 13137 | -0.002 | 0.1562 | No |
| 1769 | ATP6V1E2 | ATP6V1E2 | 13144 | -0.002 | 0.1559 | No |
| 1770 | LRGUK | LRGUK | 13146 | -0.002 | 0.1558 | No |
| 1771 | ANKZF1 | ANKZF1 | 13199 | -0.003 | 0.1532 | No |
| 1772 | SNX13 | SNX13 | 13228 | -0.005 | 0.1518 | No |
| 1773 | CKB | CKB | 13233 | -0.005 | 0.1516 | No |
| 1774 | DENND4B | DENND4B | 13234 | -0.005 | 0.1516 | No |
| 1775 | ABHD17A | ABHD17A | 13270 | -0.007 | 0.1498 | No |
| 1776 | IRF2BP1 | IRF2BP1 | 13276 | -0.008 | 0.1496 | No |
| 1777 | GPRC5B | GPRC5B | 13298 | -0.009 | 0.1485 | No |
| 1778 | RAB24 | RAB24 | 13312 | -0.011 | 0.1479 | No |
| 1779 | SNRNP25 | SNRNP25 | 13328 | -0.012 | 0.1471 | No |
| 1780 | EZH1 | EZH1 | 13341 | -0.013 | 0.1465 | No |
| 1781 | R3HDM2 | R3HDM2 | 13363 | -0.014 | 0.1455 | No |
| 1782 | DAAM1 | DAAM1 | 13370 | -0.014 | 0.1452 | No |
| 1783 | KAT5 | KAT5 | 13371 | -0.015 | 0.1452 | No |
| 1784 | CCDC130 | CCDC130 | 13385 | -0.015 | 0.1445 | No |
| 1785 | PRRG2 | PRRG2 | 13399 | -0.017 | 0.1439 | No |
| 1786 | TMEM169 | TMEM169 | 13405 | -0.017 | 0.1436 | No |
| 1787 | MCUR1 | MCUR1 | 13406 | -0.017 | 0.1436 | No |
| 1788 | TMEM218 | TMEM218 | 13410 | -0.018 | 0.1435 | No |
| 1789 | THYN1 | THYN1 | 13421 | -0.018 | 0.1430 | No |
| 1790 | TRIM28 | TRIM28 | 13456 | -0.021 | 0.1413 | No |
| 1791 | MRPS14 | MRPS14 | 13481 | -0.023 | 0.1401 | No |
| 1792 | KLHL8 | KLHL8 | 13512 | -0.025 | 0.1386 | No |
| 1793 | RAB3IP | RAB3IP | 13514 | -0.025 | 0.1386 | No |
| 1794 | RBBP4 | RBBP4 | 13524 | -0.025 | 0.1381 | No |
| 1795 | ABCB10 | ABCB10 | 13528 | -0.025 | 0.1380 | No |
| 1796 | PEG3 | PEG3 | 13563 | -0.028 | 0.1363 | No |
| 1797 | PRKCE | PRKCE | 13587 | -0.030 | 0.1351 | No |
| 1798 | MYOM1 | MYOM1 | 13593 | -0.031 | 0.1349 | No |
| 1799 | SOCS7 | SOCS7 | 13609 | -0.031 | 0.1342 | No |
| 1800 | MAZ | MAZ | 13612 | -0.032 | 0.1341 | No |
| 1801 | CLCC1 | CLCC1 | 13622 | -0.033 | 0.1336 | No |
| 1802 | E2F6 | E2F6 | 13638 | -0.034 | 0.1329 | No |
| 1803 | PPT1 | PPT1 | 13654 | -0.035 | 0.1322 | No |
| 1804 | RBM8A | RBM8A | 13657 | -0.036 | 0.1321 | No |
| 1805 | MBOAT2 | MBOAT2 | 13658 | -0.036 | 0.1321 | No |
| 1806 | PLEKHH2 | PLEKHH2 | 13664 | -0.036 | 0.1319 | No |
| 1807 | DUSP8 | DUSP8 | 13665 | -0.036 | 0.1319 | No |
| 1808 | TAF6L | TAF6L | 13672 | -0.037 | 0.1316 | No |
| 1809 | TERF2 | TERF2 | 13686 | -0.038 | 0.1310 | No |
| 1810 | MYO9B | MYO9B | 13687 | -0.038 | 0.1310 | No |
| 1811 | RBFA | RBFA | 13692 | -0.038 | 0.1308 | No |
| 1812 | ATP11A | ATP11A | 13696 | -0.038 | 0.1307 | No |
| 1813 | ELAVL1 | ELAVL1 | 13699 | -0.039 | 0.1306 | No |
| 1814 | NKIRAS1 | NKIRAS1 | 13705 | -0.039 | 0.1304 | No |
| 1815 | LSM7 | LSM7 | 13711 | -0.040 | 0.1302 | No |
| 1816 | ERI3 | ERI3 | 13730 | -0.041 | 0.1293 | No |
| 1817 | MIER3 | MIER3 | 13735 | -0.041 | 0.1291 | No |
| 1818 | SIPA1L1 | SIPA1L1 | 13748 | -0.043 | 0.1285 | No |
| 1819 | GPATCH2L | GPATCH2L | 13756 | -0.043 | 0.1282 | No |
| 1820 | ANKHD1 | ANKHD1 | 13771 | -0.044 | 0.1275 | No |
| 1821 | TIGAR | TIGAR | 13802 | -0.046 | 0.1260 | No |
| 1822 | S100PBP | S100PBP | 13832 | -0.048 | 0.1246 | No |
| 1823 | AAAS | AAAS | 13833 | -0.048 | 0.1246 | No |
| 1824 | RAC3 | RAC3 | 13853 | -0.050 | 0.1237 | No |
| 1825 | PLEKHJ1 | PLEKHJ1 | 13867 | -0.050 | 0.1230 | No |
| 1826 | AXIN1 | AXIN1 | 13886 | -0.052 | 0.1222 | No |
| 1827 | MAPK1 | MAPK1 | 13887 | -0.052 | 0.1222 | No |
| 1828 | RALGAPA1 | RALGAPA1 | 13888 | -0.052 | 0.1222 | No |
| 1829 | ZMYND19 | ZMYND19 | 13890 | -0.053 | 0.1222 | No |
| 1830 | TXN2 | TXN2 | 13893 | -0.053 | 0.1221 | No |
| 1831 | KRI1 | KRI1 | 13900 | -0.053 | 0.1219 | No |
| 1832 | DGKZ | DGKZ | 13907 | -0.054 | 0.1216 | No |
| 1833 | KAT8 | KAT8 | 13914 | -0.054 | 0.1213 | No |
| 1834 | LMO1 | LMO1 | 13915 | -0.054 | 0.1214 | No |
| 1835 | SPHK2 | SPHK2 | 13921 | -0.054 | 0.1211 | No |
| 1836 | APH1A | APH1A | 13922 | -0.054 | 0.1212 | No |
| 1837 | FAM117B | FAM117B | 13926 | -0.055 | 0.1211 | No |
| 1838 | RNMT | RNMT | 13928 | -0.055 | 0.1210 | No |
| 1839 | EIF4ENIF1 | EIF4ENIF1 | 13931 | -0.055 | 0.1210 | No |
| 1840 | AGPAT5 | AGPAT5 | 13944 | -0.055 | 0.1204 | No |
| 1841 | PRMT1 | PRMT1 | 13950 | -0.056 | 0.1202 | No |
| 1842 | GNAZ | GNAZ | 13951 | -0.056 | 0.1202 | No |
| 1843 | ZYG11B | ZYG11B | 13957 | -0.057 | 0.1200 | No |
| 1844 | NDUFA7 | NDUFA7 | 13963 | -0.057 | 0.1198 | No |
| 1845 | WSCD1 | WSCD1 | 13999 | -0.060 | 0.1180 | No |
| 1846 | PJA2 | PJA2 | 14004 | -0.061 | 0.1179 | No |
| 1847 | NFIB | NFIB | 14006 | -0.061 | 0.1179 | No |
| 1848 | HAUS8 | HAUS8 | 14030 | -0.063 | 0.1167 | No |
| 1849 | PNKP | PNKP | 14036 | -0.063 | 0.1165 | No |
| 1850 | TMEM231 | TMEM231 | 14038 | -0.063 | 0.1165 | No |
| 1851 | TAPT1 | TAPT1 | 14043 | -0.064 | 0.1163 | No |
| 1852 | LSM8 | LSM8 | 14046 | -0.064 | 0.1163 | No |
| 1853 | DHTKD1 | DHTKD1 | 14055 | -0.065 | 0.1159 | No |
| 1854 | KAT7 | KAT7 | 14066 | -0.065 | 0.1155 | No |
| 1855 | TNRC18 | TNRC18 | 14068 | -0.065 | 0.1154 | No |
| 1856 | SLC26A11 | SLC26A11 | 14075 | -0.066 | 0.1152 | No |
| 1857 | HINT3 | HINT3 | 14078 | -0.066 | 0.1151 | No |
| 1858 | GSK3A | GSK3A | 14080 | -0.066 | 0.1151 | No |
| 1859 | EIF2B5 | EIF2B5 | 14081 | -0.066 | 0.1151 | No |
| 1860 | CCDC15 | CCDC15 | 14088 | -0.066 | 0.1149 | No |
| 1861 | NRF1 | NRF1 | 14095 | -0.067 | 0.1146 | No |
| 1862 | PDCD2L | PDCD2L | 14101 | -0.067 | 0.1144 | No |
| 1863 | FOXRED2 | FOXRED2 | 14108 | -0.068 | 0.1141 | No |
| 1864 | ARMC8 | ARMC8 | 14111 | -0.068 | 0.1141 | No |
| 1865 | UNC119 | UNC119 | 14112 | -0.068 | 0.1141 | No |
| 1866 | PHACTR4 | PHACTR4 | 14117 | -0.068 | 0.1140 | No |
| 1867 | MAN2A2 | MAN2A2 | 14127 | -0.069 | 0.1136 | No |
| 1868 | TRMT5 | TRMT5 | 14129 | -0.069 | 0.1135 | No |
| 1869 | CCDC151 | CCDC151 | 14157 | -0.072 | 0.1122 | No |
| 1870 | CS | CS | 14161 | -0.072 | 0.1121 | No |
| 1871 | ZC3H10 | ZC3H10 | 14167 | -0.072 | 0.1119 | No |
| 1872 | TM2D3 | TM2D3 | 14169 | -0.072 | 0.1119 | No |
| 1873 | PLCXD2 | PLCXD2 | 14192 | -0.075 | 0.1108 | No |
| 1874 | MARC2 | MARC2 | 14207 | -0.076 | 0.1102 | No |
| 1875 | ENOPH1 | ENOPH1 | 14215 | -0.076 | 0.1099 | No |
| 1876 | TIMM44 | TIMM44 | 14218 | -0.076 | 0.1098 | No |
| 1877 | TTPA | TTPA | 14220 | -0.076 | 0.1098 | No |
| 1878 | GRK4 | GRK4 | 14224 | -0.076 | 0.1097 | No |
| 1879 | SDHAF4 | SDHAF4 | 14251 | -0.079 | 0.1084 | No |
| 1880 | YES1 | YES1 | 14252 | -0.079 | 0.1085 | No |
| 1881 | SEPT7 | SEPT7 | 14260 | -0.079 | 0.1082 | No |
| 1882 | PARS2 | PARS2 | 14267 | -0.080 | 0.1079 | No |
| 1883 | GREB1L | GREB1L | 14269 | -0.080 | 0.1079 | No |
| 1884 | BBOX1 | BBOX1 | 14283 | -0.081 | 0.1073 | No |
| 1885 | PLEKHO1 | PLEKHO1 | 14299 | -0.082 | 0.1066 | No |
| 1886 | RAPGEF2 | RAPGEF2 | 14308 | -0.082 | 0.1062 | No |
| 1887 | SARS2 | SARS2 | 14314 | -0.082 | 0.1060 | No |
| 1888 | MED28 | MED28 | 14325 | -0.083 | 0.1056 | No |
| 1889 | JDP2 | JDP2 | 14334 | -0.083 | 0.1052 | No |
| 1890 | PCYOX1L | PCYOX1L | 14337 | -0.084 | 0.1052 | No |
| 1891 | CACNA1I | CACNA1I | 14340 | -0.084 | 0.1051 | No |
| 1892 | GK | GK | 14350 | -0.085 | 0.1047 | No |
| 1893 | TAOK3 | TAOK3 | 14359 | -0.085 | 0.1044 | No |
| 1894 | ABHD14A | ABHD14A | 14372 | -0.086 | 0.1038 | No |
| 1895 | SLC41A1 | SLC41A1 | 14376 | -0.086 | 0.1037 | No |
| 1896 | NNT | NNT | 14399 | -0.088 | 0.1027 | No |
| 1897 | SFXN1 | SFXN1 | 14409 | -0.088 | 0.1023 | No |
| 1898 | RHOQ | RHOQ | 14433 | -0.090 | 0.1011 | No |
| 1899 | OBSL1 | OBSL1 | 14443 | -0.091 | 0.1007 | No |
| 1900 | NRN1L | NRN1L | 14457 | -0.092 | 0.1001 | No |
| 1901 | RAB3GAP2 | RAB3GAP2 | 14464 | -0.092 | 0.0999 | No |
| 1902 | CBX3 | CBX3 | 14468 | -0.093 | 0.0998 | No |
| 1903 | GAB2 | GAB2 | 14479 | -0.094 | 0.0994 | No |
| 1904 | FBXW8 | FBXW8 | 14494 | -0.094 | 0.0987 | No |
| 1905 | CPT2 | CPT2 | 14496 | -0.094 | 0.0987 | No |
| 1906 | ERCC3 | ERCC3 | 14507 | -0.095 | 0.0983 | No |
| 1907 | AKT2 | AKT2 | 14517 | -0.096 | 0.0979 | No |
| 1908 | STAU2 | STAU2 | 14526 | -0.097 | 0.0975 | No |
| 1909 | COG7 | COG7 | 14531 | -0.097 | 0.0974 | No |
| 1910 | SAMD1 | SAMD1 | 14537 | -0.098 | 0.0972 | No |
| 1911 | USP30 | USP30 | 14549 | -0.099 | 0.0967 | No |
| 1912 | COPG2 | COPG2 | 14572 | -0.102 | 0.0956 | No |
| 1913 | ETV1 | ETV1 | 14589 | -0.103 | 0.0949 | No |
| 1914 | DPY19L1 | DPY19L1 | 14597 | -0.104 | 0.0946 | No |
| 1915 | TMEM183A | TMEM183A | 14611 | -0.105 | 0.0940 | No |
| 1916 | MIB1 | MIB1 | 14620 | -0.106 | 0.0937 | No |
| 1917 | ATP5B | ATP5B | 14629 | -0.107 | 0.0933 | No |
| 1918 | NR6A1 | NR6A1 | 14633 | -0.107 | 0.0932 | No |
| 1919 | PANK1 | PANK1 | 14641 | -0.108 | 0.0929 | No |
| 1920 | CTNNBIP1 | CTNNBIP1 | 14656 | -0.109 | 0.0923 | No |
| 1921 | DDX51 | DDX51 | 14688 | -0.111 | 0.0908 | No |
| 1922 | SEPT8 | SEPT8 | 14692 | -0.111 | 0.0907 | No |
| 1923 | ARL10 | ARL10 | 14701 | -0.112 | 0.0904 | No |
| 1924 | PASK | PASK | 14712 | -0.112 | 0.0899 | No |
| 1925 | LYRM9 | LYRM9 | 14713 | -0.113 | 0.0900 | No |
| 1926 | AGPAT4 | AGPAT4 | 14734 | -0.115 | 0.0891 | No |
| 1927 | FAM184A | FAM184A | 14739 | -0.115 | 0.0889 | No |
| 1928 | CPT1C | CPT1C | 14762 | -0.116 | 0.0879 | No |
| 1929 | DDX49 | DDX49 | 14767 | -0.117 | 0.0878 | No |
| 1930 | SKP2 | SKP2 | 14777 | -0.118 | 0.0874 | No |
| 1931 | IGSF3 | IGSF3 | 14787 | -0.119 | 0.0870 | No |
| 1932 | TMEM128 | TMEM128 | 14815 | -0.121 | 0.0857 | No |
| 1933 | IDH2 | IDH2 | 14826 | -0.122 | 0.0853 | No |
| 1934 | SCARNA2 | SCARNA2 | 14830 | -0.123 | 0.0852 | No |
| 1935 | TYMS | TYMS | 14837 | -0.123 | 0.0850 | No |
| 1936 | RBM22 | RBM22 | 14840 | -0.124 | 0.0849 | No |
| 1937 | HHATL | HHATL | 14847 | -0.124 | 0.0847 | No |
| 1938 | BAIAP3 | BAIAP3 | 14852 | -0.124 | 0.0846 | No |
| 1939 | TSPAN33 | TSPAN33 | 14878 | -0.124 | 0.0834 | No |
| 1940 | VARS2 | VARS2 | 14880 | -0.124 | 0.0834 | No |
| 1941 | PGAP3 | PGAP3 | 14882 | -0.125 | 0.0835 | No |
| 1942 | ACSL6 | ACSL6 | 14885 | -0.125 | 0.0834 | No |
| 1943 | TEKT1 | TEKT1 | 14888 | -0.125 | 0.0834 | No |
| 1944 | TRPM3 | TRPM3 | 14892 | -0.125 | 0.0833 | No |
| 1945 | IFNAR2 | IFNAR2 | 14907 | -0.125 | 0.0827 | No |
| 1946 | EPN2 | EPN2 | 14917 | -0.126 | 0.0823 | No |
| 1947 | ATP5D | ATP5D | 14919 | -0.126 | 0.0823 | No |
| 1948 | CLPB | CLPB | 14931 | -0.127 | 0.0819 | No |
| 1949 | ATP2A2 | ATP2A2 | 14939 | -0.128 | 0.0816 | No |
| 1950 | LRRC58 | LRRC58 | 14940 | -0.128 | 0.0817 | No |
| 1951 | SEC11C | SEC11C | 14962 | -0.130 | 0.0807 | No |
| 1952 | P2RY1 | P2RY1 | 14963 | -0.130 | 0.0808 | No |
| 1953 | APP | APP | 14965 | -0.130 | 0.0808 | No |
| 1954 | RAVER2 | RAVER2 | 14977 | -0.131 | 0.0803 | No |
| 1955 | UCP3 | UCP3 | 14994 | -0.133 | 0.0796 | No |
| 1956 | SCARNA17 | SCARNA17 | 15001 | -0.133 | 0.0794 | No |
| 1957 | DMTN | DMTN | 15027 | -0.134 | 0.0782 | No |
| 1958 | RYR1 | RYR1 | 15029 | -0.134 | 0.0782 | No |
| 1959 | PRMT7 | PRMT7 | 15088 | -0.138 | 0.0754 | No |
| 1960 | GLCCI1 | GLCCI1 | 15122 | -0.142 | 0.0738 | No |
| 1961 | GFM1 | GFM1 | 15152 | -0.145 | 0.0724 | No |
| 1962 | RHBDD2 | RHBDD2 | 15168 | -0.146 | 0.0717 | No |
| 1963 | LANCL2 | LANCL2 | 15175 | -0.147 | 0.0715 | No |
| 1964 | ATP6V0A2 | ATP6V0A2 | 15179 | -0.147 | 0.0714 | No |
| 1965 | SLC25A15 | SLC25A15 | 15181 | -0.147 | 0.0715 | No |
| 1966 | KANSL1L | KANSL1L | 15196 | -0.148 | 0.0709 | No |
| 1967 | NYAP1 | NYAP1 | 15204 | -0.149 | 0.0706 | No |
| 1968 | SLC36A1 | SLC36A1 | 15213 | -0.150 | 0.0703 | No |
| 1969 | BCKDHA | BCKDHA | 15220 | -0.150 | 0.0701 | No |
| 1970 | GTPBP1 | GTPBP1 | 15223 | -0.151 | 0.0701 | No |
| 1971 | PRTG | PRTG | 15225 | -0.151 | 0.0701 | No |
| 1972 | SEMA6D | SEMA6D | 15241 | -0.153 | 0.0694 | No |
| 1973 | CC2D1A | CC2D1A | 15249 | -0.154 | 0.0692 | No |
| 1974 | ABHD3 | ABHD3 | 15286 | -0.157 | 0.0675 | No |
| 1975 | PIGF | PIGF | 15295 | -0.158 | 0.0671 | No |
| 1976 | CFAP36 | CFAP36 | 15335 | -0.162 | 0.0653 | No |
| 1977 | C1QTNF4 | C1QTNF4 | 15346 | -0.163 | 0.0649 | No |
| 1978 | ZBTB10 | ZBTB10 | 15354 | -0.164 | 0.0646 | No |
| 1979 | TLK1 | TLK1 | 15374 | -0.165 | 0.0637 | No |
| 1980 | PTPN4 | PTPN4 | 15377 | -0.165 | 0.0637 | No |
| 1981 | MEGF9 | MEGF9 | 15387 | -0.167 | 0.0634 | No |
| 1982 | UBA2 | UBA2 | 15388 | -0.167 | 0.0635 | No |
| 1983 | CDC14B | CDC14B | 15418 | -0.171 | 0.0621 | No |
| 1984 | RNF157 | RNF157 | 15434 | -0.172 | 0.0615 | No |
| 1985 | TRAP1 | TRAP1 | 15448 | -0.174 | 0.0609 | No |
| 1986 | KCTD15 | KCTD15 | 15456 | -0.175 | 0.0607 | No |
| 1987 | MDH2 | MDH2 | 15469 | -0.176 | 0.0602 | No |
| 1988 | RPRD1A | RPRD1A | 15484 | -0.178 | 0.0596 | No |
| 1989 | ZGRF1 | ZGRF1 | 15486 | -0.178 | 0.0596 | No |
| 1990 | KIF5C | KIF5C | 15503 | -0.179 | 0.0589 | No |
| 1991 | ACTR6 | ACTR6 | 15523 | -0.181 | 0.0581 | No |
| 1992 | B3GLCT | B3GLCT | 15530 | -0.182 | 0.0579 | No |
| 1993 | MUM1 | MUM1 | 15531 | -0.182 | 0.0580 | No |
| 1994 | LRRC75A | LRRC75A | 15553 | -0.184 | 0.0571 | No |
| 1995 | ASNA1 | ASNA1 | 15554 | -0.184 | 0.0572 | No |
| 1996 | LAPTM4B | LAPTM4B | 15567 | -0.187 | 0.0567 | No |
| 1997 | ZCCHC2 | ZCCHC2 | 15568 | -0.187 | 0.0568 | No |
| 1998 | LBX1 | LBX1 | 15592 | -0.189 | 0.0557 | No |
| 1999 | FGF8 | FGF8 | 15593 | -0.189 | 0.0559 | No |
| 2000 | CELF5 | CELF5 | 15607 | -0.190 | 0.0553 | No |
| 2001 | JUND | JUND | 15638 | -0.193 | 0.0539 | No |
| 2002 | KLHDC1 | KLHDC1 | 15664 | -0.195 | 0.0528 | No |
| 2003 | NDUFA10 | NDUFA10 | 15687 | -0.197 | 0.0518 | No |
| 2004 | COX18 | COX18 | 15696 | -0.198 | 0.0515 | No |
| 2005 | PSIP1 | PSIP1 | 15701 | -0.199 | 0.0514 | No |
| 2006 | TCF3 | TCF3 | 15709 | -0.200 | 0.0512 | No |
| 2007 | ALKBH7 | ALKBH7 | 15715 | -0.201 | 0.0510 | No |
| 2008 | BCL2 | BCL2 | 15716 | -0.201 | 0.0512 | No |
| 2009 | TMEM199 | TMEM199 | 15723 | -0.201 | 0.0510 | No |
| 2010 | CIAPIN1 | CIAPIN1 | 15726 | -0.202 | 0.0510 | No |
| 2011 | DSEL | DSEL | 15758 | -0.207 | 0.0496 | No |
| 2012 | PRCP | PRCP | 15761 | -0.208 | 0.0496 | No |
| 2013 | PROX1 | PROX1 | 15766 | -0.208 | 0.0495 | No |
| 2014 | NCK1 | NCK1 | 15797 | -0.212 | 0.0481 | No |
| 2015 | PTH1R | PTH1R | 15799 | -0.213 | 0.0482 | No |
| 2016 | DOCK6 | DOCK6 | 15810 | -0.213 | 0.0478 | No |
| 2017 | AP1S1 | AP1S1 | 15814 | -0.214 | 0.0478 | No |
| 2018 | TJP1 | TJP1 | 15818 | -0.214 | 0.0478 | No |
| 2019 | COL20A1 | COL20A1 | 15846 | -0.217 | 0.0465 | No |
| 2020 | PHYH | PHYH | 15858 | -0.218 | 0.0461 | No |
| 2021 | HOMER1 | HOMER1 | 15906 | -0.224 | 0.0439 | No |
| 2022 | USP42 | USP42 | 15907 | -0.224 | 0.0440 | No |
| 2023 | HOPX | HOPX | 15911 | -0.225 | 0.0440 | No |
| 2024 | STIM2 | STIM2 | 15923 | -0.226 | 0.0436 | No |
| 2025 | BEND7 | BEND7 | 15934 | -0.227 | 0.0432 | No |
| 2026 | SYT17 | SYT17 | 15937 | -0.228 | 0.0433 | No |
| 2027 | FRAT2 | FRAT2 | 15938 | -0.228 | 0.0434 | No |
| 2028 | ANKRD49 | ANKRD49 | 15954 | -0.229 | 0.0428 | No |
| 2029 | CCL25 | CCL25 | 15963 | -0.230 | 0.0425 | No |
| 2030 | LYSMD2 | LYSMD2 | 15969 | -0.231 | 0.0424 | No |
| 2031 | FOXO6 | FOXO6 | 15970 | -0.231 | 0.0425 | No |
| 2032 | ABHD6 | ABHD6 | 15975 | -0.231 | 0.0425 | No |
| 2033 | MYH14 | MYH14 | 15984 | -0.233 | 0.0422 | No |
| 2034 | RIMS2 | RIMS2 | 15988 | -0.233 | 0.0422 | No |
| 2035 | CTXN2 | CTXN2 | 15993 | -0.233 | 0.0421 | No |
| 2036 | SEC14L1 | SEC14L1 | 16008 | -0.234 | 0.0416 | No |
| 2037 | PON3 | PON3 | 16028 | -0.235 | 0.0408 | No |
| 2038 | HAUS3 | HAUS3 | 16047 | -0.237 | 0.0400 | No |
| 2039 | THSD7A | THSD7A | 16063 | -0.240 | 0.0394 | No |
| 2040 | ADNP2 | ADNP2 | 16071 | -0.241 | 0.0392 | No |
| 2041 | BTBD3 | BTBD3 | 16081 | -0.242 | 0.0389 | No |
| 2042 | RPA2 | RPA2 | 16102 | -0.244 | 0.0380 | No |
| 2043 | ST8SIA1 | ST8SIA1 | 16103 | -0.244 | 0.0381 | No |
| 2044 | SLC48A1 | SLC48A1 | 16105 | -0.245 | 0.0382 | No |
| 2045 | TBRG4 | TBRG4 | 16120 | -0.246 | 0.0377 | No |
| 2046 | LONRF1 | LONRF1 | 16127 | -0.247 | 0.0375 | No |
| 2047 | DHPS | DHPS | 16139 | -0.248 | 0.0371 | No |
| 2048 | SLC12A5 | SLC12A5 | 16141 | -0.248 | 0.0372 | No |
| 2049 | ZFAND2B | ZFAND2B | 16158 | -0.251 | 0.0366 | No |
| 2050 | KAT2A | KAT2A | 16160 | -0.251 | 0.0367 | No |
| 2051 | FOXK1 | FOXK1 | 16161 | -0.251 | 0.0368 | No |
| 2052 | KHK | KHK | 16171 | -0.252 | 0.0365 | No |
| 2053 | RHOU | RHOU | 16186 | -0.254 | 0.0360 | No |
| 2054 | TMEM51 | TMEM51 | 16191 | -0.254 | 0.0359 | No |
| 2055 | RN7SK | RN7SK | 16196 | -0.255 | 0.0359 | No |
| 2056 | ZADH2 | ZADH2 | 16200 | -0.256 | 0.0359 | No |
| 2057 | OXR1 | OXR1 | 16203 | -0.256 | 0.0359 | No |
| 2058 | ULK3 | ULK3 | 16225 | -0.259 | 0.0350 | No |
| 2059 | INSR | INSR | 16258 | -0.262 | 0.0336 | No |
| 2060 | JAG1 | JAG1 | 16274 | -0.264 | 0.0330 | No |
| 2061 | BAG1 | BAG1 | 16306 | -0.269 | 0.0316 | No |
| 2062 | NKPD1 | NKPD1 | 16311 | -0.270 | 0.0315 | No |
| 2063 | ZGLP1 | ZGLP1 | 16312 | -0.270 | 0.0317 | No |
| 2064 | TFRC | TFRC | 16319 | -0.271 | 0.0316 | No |
| 2065 | HIP1 | HIP1 | 16324 | -0.271 | 0.0315 | No |
| 2066 | RPS6KA2 | RPS6KA2 | 16330 | -0.272 | 0.0314 | No |
| 2067 | QDPR | QDPR | 16338 | -0.274 | 0.0312 | No |
| 2068 | ACACB | ACACB | 16363 | -0.276 | 0.0302 | No |
| 2069 | GLDC | GLDC | 16388 | -0.281 | 0.0291 | No |
| 2070 | WDR18 | WDR18 | 16391 | -0.281 | 0.0292 | No |
| 2071 | ZDHHC15 | ZDHHC15 | 16402 | -0.282 | 0.0289 | No |
| 2072 | FLOT2 | FLOT2 | 16419 | -0.284 | 0.0282 | No |
| 2073 | MERTK | MERTK | 16429 | -0.285 | 0.0280 | No |
| 2074 | MOB1B | MOB1B | 16430 | -0.286 | 0.0281 | No |
| 2075 | IQGAP2 | IQGAP2 | 16431 | -0.286 | 0.0283 | No |
| 2076 | GOLM1 | GOLM1 | 16433 | -0.286 | 0.0284 | No |
| 2077 | SPATA6 | SPATA6 | 16435 | -0.287 | 0.0286 | No |
| 2078 | TMEM143 | TMEM143 | 16436 | -0.287 | 0.0287 | No |
| 2079 | UBE2QL1 | UBE2QL1 | 16446 | -0.289 | 0.0285 | No |
| 2080 | RILPL1 | RILPL1 | 16450 | -0.290 | 0.0285 | No |
| 2081 | MIIP | MIIP | 16454 | -0.290 | 0.0285 | No |
| 2082 | PSPH | PSPH | 16473 | -0.292 | 0.0278 | No |
| 2083 | PPAN | PPAN | 16495 | -0.296 | 0.0269 | No |
| 2084 | YBX1 | YBX1 | 16521 | -0.300 | 0.0258 | No |
| 2085 | ATP9B | ATP9B | 16538 | -0.302 | 0.0252 | No |
| 2086 | BBC3 | BBC3 | 16546 | -0.304 | 0.0250 | No |
| 2087 | AATK | AATK | 16548 | -0.304 | 0.0251 | No |
| 2088 | TCAP | TCAP | 16561 | -0.305 | 0.0247 | No |
| 2089 | CYP2U1 | CYP2U1 | 16571 | -0.307 | 0.0245 | No |
| 2090 | BTAF1 | BTAF1 | 16574 | -0.308 | 0.0245 | No |
| 2091 | ERF | ERF | 16578 | -0.309 | 0.0246 | No |
| 2092 | SYT4 | SYT4 | 16584 | -0.310 | 0.0245 | No |
| 2093 | NDFIP1 | NDFIP1 | 16591 | -0.311 | 0.0244 | No |
| 2094 | USF2 | USF2 | 16592 | -0.311 | 0.0246 | No |
| 2095 | CCDC184 | CCDC184 | 16597 | -0.312 | 0.0246 | No |
| 2096 | DAGLA | DAGLA | 16606 | -0.314 | 0.0244 | No |
| 2097 | TMEM50B | TMEM50B | 16640 | -0.318 | 0.0229 | No |
| 2098 | ILVBL | ILVBL | 16647 | -0.320 | 0.0228 | No |
| 2099 | FRS3 | FRS3 | 16651 | -0.321 | 0.0228 | No |
| 2100 | SMOC2 | SMOC2 | 16658 | -0.322 | 0.0227 | No |
| 2101 | FLVCR1 | FLVCR1 | 16678 | -0.324 | 0.0219 | No |
| 2102 | CAND2 | CAND2 | 16689 | -0.327 | 0.0216 | No |
| 2103 | SPSB4 | SPSB4 | 16698 | -0.328 | 0.0214 | No |
| 2104 | NCKAP1L | NCKAP1L | 16707 | -0.330 | 0.0212 | No |
| 2105 | LIG1 | LIG1 | 16715 | -0.331 | 0.0211 | No |
| 2106 | DIRAS1 | DIRAS1 | 16752 | -0.339 | 0.0195 | No |
| 2107 | UBQLN4 | UBQLN4 | 16756 | -0.339 | 0.0195 | No |
| 2108 | P2RX7 | P2RX7 | 16761 | -0.340 | 0.0195 | No |
| 2109 | CORO2B | CORO2B | 16789 | -0.346 | 0.0184 | No |
| 2110 | SCAF4 | SCAF4 | 16821 | -0.352 | 0.0170 | No |
| 2111 | PANK4 | PANK4 | 16843 | -0.354 | 0.0162 | No |
| 2112 | SHC3 | SHC3 | 16855 | -0.356 | 0.0158 | No |
| 2113 | SLC39A6 | SLC39A6 | 16884 | -0.361 | 0.0146 | No |
| 2114 | SEMA4C | SEMA4C | 16889 | -0.361 | 0.0146 | No |
| 2115 | NADK2 | NADK2 | 16891 | -0.362 | 0.0148 | No |
| 2116 | VASH1 | VASH1 | 16902 | -0.364 | 0.0145 | No |
| 2117 | IER5 | IER5 | 16913 | -0.365 | 0.0142 | No |
| 2118 | PSAT1 | PSAT1 | 16921 | -0.366 | 0.0141 | No |
| 2119 | DNAJB2 | DNAJB2 | 16926 | -0.368 | 0.0141 | No |
| 2120 | ICA1 | ICA1 | 16935 | -0.370 | 0.0140 | No |
| 2121 | TBC1D12 | TBC1D12 | 16965 | -0.375 | 0.0127 | No |
| 2122 | GINS2 | GINS2 | 16979 | -0.377 | 0.0123 | No |
| 2123 | CCSAP | CCSAP | 16983 | -0.378 | 0.0124 | No |
| 2124 | APBB2 | APBB2 | 17007 | -0.382 | 0.0114 | No |
| 2125 | EIF4EBP2 | EIF4EBP2 | 17013 | -0.383 | 0.0114 | No |
| 2126 | CDCA7 | CDCA7 | 17041 | -0.389 | 0.0103 | No |
| 2127 | CDCA7L | CDCA7L | 17050 | -0.391 | 0.0101 | No |
| 2128 | ASGR1 | ASGR1 | 17059 | -0.393 | 0.0100 | No |
| 2129 | PPP5C | PPP5C | 17060 | -0.393 | 0.0102 | No |
| 2130 | CCND2 | CCND2 | 17068 | -0.395 | 0.0101 | No |
| 2131 | ANKRD12 | ANKRD12 | 17072 | -0.396 | 0.0102 | No |
| 2132 | IL1RAP | IL1RAP | 17145 | -0.407 | 0.0068 | No |
| 2133 | TMOD4 | TMOD4 | 17159 | -0.411 | 0.0064 | No |
| 2134 | NR2C2AP | NR2C2AP | 17164 | -0.411 | 0.0064 | No |
| 2135 | TMEM251 | TMEM251 | 17178 | -0.414 | 0.0060 | No |
| 2136 | LYRM7 | LYRM7 | 17200 | -0.417 | 0.0052 | No |
| 2137 | TNKS | TNKS | 17208 | -0.418 | 0.0051 | No |
| 2138 | PLEKHH3 | PLEKHH3 | 17227 | -0.422 | 0.0044 | No |
| 2139 | URB2 | URB2 | 17238 | -0.426 | 0.0042 | No |
| 2140 | AES | AES | 17240 | -0.426 | 0.0044 | No |
| 2141 | NGB | NGB | 17248 | -0.428 | 0.0043 | No |
| 2142 | PUSL1 | PUSL1 | 17250 | -0.428 | 0.0045 | No |
| 2143 | NUDT6 | NUDT6 | 17252 | -0.428 | 0.0047 | No |
| 2144 | LPIN2 | LPIN2 | 17257 | -0.429 | 0.0048 | No |
| 2145 | JADE3 | JADE3 | 17259 | -0.430 | 0.0050 | No |
| 2146 | KCTD13 | KCTD13 | 17264 | -0.431 | 0.0051 | No |
| 2147 | FAM3C | FAM3C | 17285 | -0.436 | 0.0043 | No |
| 2148 | STK32B | STK32B | 17297 | -0.438 | 0.0040 | No |
| 2149 | TMEM120A | TMEM120A | 17299 | -0.439 | 0.0042 | No |
| 2150 | GAS1 | GAS1 | 17305 | -0.440 | 0.0043 | No |
| 2151 | TBR1 | TBR1 | 17318 | -0.444 | 0.0039 | No |
| 2152 | RIMKLA | RIMKLA | 17325 | -0.445 | 0.0039 | No |
| 2153 | KLHL28 | KLHL28 | 17326 | -0.445 | 0.0042 | No |
| 2154 | ST3GAL3 | ST3GAL3 | 17327 | -0.445 | 0.0044 | No |
| 2155 | ITPR2 | ITPR2 | 17333 | -0.446 | 0.0045 | No |
| 2156 | TIMELESS | TIMELESS | 17352 | -0.449 | 0.0038 | No |
| 2157 | ZYX | ZYX | 17383 | -0.458 | 0.0026 | No |
| 2158 | FLRT1 | FLRT1 | 17402 | -0.462 | 0.0019 | No |
| 2159 | UBE2G2 | UBE2G2 | 17412 | -0.464 | 0.0018 | No |
| 2160 | ETV5 | ETV5 | 17415 | -0.464 | 0.0020 | No |
| 2161 | GRASP | GRASP | 17427 | -0.467 | 0.0017 | No |
| 2162 | AMIGO1 | AMIGO1 | 17432 | -0.468 | 0.0018 | No |
| 2163 | RIMS3 | RIMS3 | 17433 | -0.468 | 0.0020 | No |
| 2164 | SLC4A8 | SLC4A8 | 17443 | -0.470 | 0.0019 | No |
| 2165 | NEURL1B | NEURL1B | 17464 | -0.475 | 0.0012 | No |
| 2166 | KCNQ4 | KCNQ4 | 17481 | -0.481 | 0.0006 | No |
| 2167 | SH2D3C | SH2D3C | 17496 | -0.485 | 0.0002 | No |
| 2168 | ZSWIM7 | ZSWIM7 | 17503 | -0.487 | 0.0002 | No |
| 2169 | INHA | INHA | 17528 | -0.496 | -0.0007 | No |
| 2170 | MAP6 | MAP6 | 17544 | -0.499 | -0.0011 | No |
| 2171 | CELF4 | CELF4 | 17564 | -0.505 | -0.0018 | No |
| 2172 | CCDC97 | CCDC97 | 17578 | -0.510 | -0.0022 | No |
| 2173 | SFT2D3 | SFT2D3 | 17587 | -0.513 | -0.0022 | No |
| 2174 | LEPROTL1 | LEPROTL1 | 17588 | -0.513 | -0.0019 | No |
| 2175 | ARHGEF2 | ARHGEF2 | 17601 | -0.515 | -0.0022 | No |
| 2176 | CEBPG | CEBPG | 17623 | -0.522 | -0.0030 | No |
| 2177 | LRRC27 | LRRC27 | 17647 | -0.530 | -0.0038 | No |
| 2178 | ITPRIPL1 | ITPRIPL1 | 17653 | -0.533 | -0.0037 | No |
| 2179 | NUDT12 | NUDT12 | 17657 | -0.534 | -0.0036 | No |
| 2180 | VPS13C | VPS13C | 17682 | -0.539 | -0.0045 | No |
| 2181 | SLC25A33 | SLC25A33 | 17693 | -0.542 | -0.0046 | No |
| 2182 | WDR62 | WDR62 | 17729 | -0.559 | -0.0061 | No |
| 2183 | PDPR | PDPR | 17801 | -0.585 | -0.0093 | No |
| 2184 | STAC3 | STAC3 | 17802 | -0.585 | -0.0089 | No |
| 2185 | RNASE6 | RNASE6 | 17831 | -0.596 | -0.0100 | No |
| 2186 | EFNA1 | EFNA1 | 17837 | -0.597 | -0.0099 | No |
| 2187 | PTPN13 | PTPN13 | 17838 | -0.597 | -0.0095 | No |
| 2188 | CLDN15 | CLDN15 | 17844 | -0.599 | -0.0094 | No |
| 2189 | SLC5A1 | SLC5A1 | 17851 | -0.599 | -0.0093 | No |
| 2190 | MYH3 | MYH3 | 17867 | -0.599 | -0.0097 | No |
| 2191 | TERT | TERT | 17872 | -0.599 | -0.0096 | No |
| 2192 | GRIK1 | GRIK1 | 17878 | -0.599 | -0.0095 | No |
| 2193 | ATRIP | ATRIP | 17890 | -0.599 | -0.0097 | No |
| 2194 | SLC11A1 | SLC11A1 | 17892 | -0.599 | -0.0093 | No |
| 2195 | RPE65 | RPE65 | 17904 | -0.599 | -0.0095 | No |
| 2196 | TTC21A | TTC21A | 17936 | -0.599 | -0.0107 | No |
| 2197 | CPNE4 | CPNE4 | 17937 | -0.599 | -0.0104 | No |
| 2198 | HSF4 | HSF4 | 17941 | -0.599 | -0.0102 | No |
| 2199 | BARHL2 | BARHL2 | 17943 | -0.599 | -0.0098 | No |
| 2200 | ZMAT4 | ZMAT4 | 17955 | -0.599 | -0.0100 | No |
| 2201 | TG | TG | 17997 | -0.599 | -0.0117 | No |
| 2202 | RDH16 | RDH16 | 18059 | -0.599 | -0.0145 | No |
| 2203 | CCDC78 | CCDC78 | 18063 | -0.599 | -0.0143 | No |
| 2204 | SNORA47 | SNORA47 | 18198 | -0.599 | -0.0207 | No |
| 2205 | CHRD | CHRD | 18399 | -0.600 | -0.0304 | No |
| 2206 | TBX21 | TBX21 | 18408 | -0.602 | -0.0305 | No |
| 2207 | ATP1B4 | ATP1B4 | 18418 | -0.602 | -0.0306 | No |
| 2208 | WFDC2 | WFDC2 | 18420 | -0.602 | -0.0302 | No |
| 2209 | BPIFB1 | BPIFB1 | 18455 | -0.602 | -0.0316 | No |
| 2210 | SLC6A12 | SLC6A12 | 18469 | -0.602 | -0.0319 | No |
| 2211 | GIPR | GIPR | 18472 | -0.602 | -0.0316 | No |
| 2212 | MYRFL | MYRFL | 18488 | -0.602 | -0.0320 | No |
| 2213 | LEAP2 | LEAP2 | 18499 | -0.602 | -0.0322 | No |
| 2214 | PRR32 | PRR32 | 18501 | -0.602 | -0.0318 | No |
| 2215 | BCL2L15 | BCL2L15 | 18520 | -0.602 | -0.0324 | No |
| 2216 | SAMD13 | SAMD13 | 18534 | -0.602 | -0.0327 | No |
| 2217 | C1QTNF3 | C1QTNF3 | 18557 | -0.602 | -0.0334 | No |
| 2218 | SNORD4A | SNORD4A | 18617 | -0.602 | -0.0360 | No |
| 2219 | CD101 | CD101 | 18726 | -0.602 | -0.0411 | No |
| 2220 | GIMAP1 | GIMAP1 | 18746 | -0.602 | -0.0417 | No |
| 2221 | PDE3B | PDE3B | 18956 | -0.604 | -0.0519 | No |
| 2222 | EGR1 | EGR1 | 18958 | -0.605 | -0.0516 | No |
| 2223 | CREB5 | CREB5 | 18961 | -0.605 | -0.0514 | No |
| 2224 | MAPKAPK5 | MAPKAPK5 | 18985 | -0.612 | -0.0521 | No |
| 2225 | CACNA2D2 | CACNA2D2 | 19014 | -0.621 | -0.0532 | No |
| 2226 | SLC18A1 | SLC18A1 | 19026 | -0.624 | -0.0534 | No |
| 2227 | KLF1 | KLF1 | 19027 | -0.624 | -0.0530 | No |
| 2228 | FAM49A | FAM49A | 19043 | -0.625 | -0.0534 | No |
| 2229 | PRPH | PRPH | 19102 | -0.630 | -0.0559 | No |
| 2230 | KCNK7 | KCNK7 | 19104 | -0.630 | -0.0556 | No |
| 2231 | CD72 | CD72 | 19111 | -0.630 | -0.0555 | No |
| 2232 | BAALC | BAALC | 19219 | -0.632 | -0.0605 | No |
| 2233 | SLA | SLA | 19220 | -0.632 | -0.0601 | No |
| 2234 | ENDOU | ENDOU | 19221 | -0.632 | -0.0598 | No |
| 2235 | GLI1 | GLI1 | 19222 | -0.632 | -0.0594 | No |
| 2236 | CALCB | CALCB | 19226 | -0.632 | -0.0591 | No |
| 2237 | CEP85L | CEP85L | 19229 | -0.632 | -0.0589 | No |
| 2238 | IGFALS | IGFALS | 19239 | -0.632 | -0.0589 | No |
| 2239 | NSL1 | NSL1 | 19317 | -0.632 | -0.0624 | No |
| 2240 | CPNE2 | CPNE2 | 19325 | -0.636 | -0.0624 | No |
| 2241 | VEGFA | VEGFA | 19327 | -0.638 | -0.0621 | No |
| 2242 | SNORD87 | SNORD87 | 19336 | -0.640 | -0.0621 | No |
| 2243 | IL12A | IL12A | 19347 | -0.642 | -0.0622 | No |
| 2244 | CLVS1 | CLVS1 | 19357 | -0.642 | -0.0623 | No |
| 2245 | OPN1SW | OPN1SW | 19365 | -0.642 | -0.0622 | No |
| 2246 | HMX2 | HMX2 | 19403 | -0.643 | -0.0637 | No |
| 2247 | HIST1H1E | HIST1H1E | 19432 | -0.647 | -0.0647 | No |
| 2248 | PRKCB | PRKCB | 19433 | -0.647 | -0.0643 | No |
| 2249 | ART3 | ART3 | 19435 | -0.648 | -0.0640 | No |
| 2250 | KCNJ4 | KCNJ4 | 19483 | -0.653 | -0.0660 | No |
| 2251 | MPP6 | MPP6 | 19518 | -0.659 | -0.0673 | No |
| 2252 | KCNC1 | KCNC1 | 19526 | -0.662 | -0.0672 | No |
| 2253 | ERBB3 | ERBB3 | 19530 | -0.663 | -0.0670 | No |
| 2254 | SLC4A10 | SLC4A10 | 19551 | -0.670 | -0.0676 | No |
| 2255 | PPARGC1B | PPARGC1B | 19570 | -0.675 | -0.0681 | No |
| 2256 | SDK2 | SDK2 | 19575 | -0.677 | -0.0679 | No |
| 2257 | ING5 | ING5 | 19598 | -0.682 | -0.0686 | No |
| 2258 | PDGFA | PDGFA | 19619 | -0.685 | -0.0692 | No |
| 2259 | PTCD3 | PTCD3 | 19652 | -0.688 | -0.0704 | No |
| 2260 | HEATR5B | HEATR5B | 19660 | -0.690 | -0.0703 | No |
| 2261 | GREB1 | GREB1 | 19695 | -0.703 | -0.0716 | No |
| 2262 | APOM | APOM | 19711 | -0.709 | -0.0719 | No |
| 2263 | ATP2B2 | ATP2B2 | 19742 | -0.709 | -0.0730 | No |
| 2264 | CABP1 | CABP1 | 19750 | -0.710 | -0.0729 | No |
| 2265 | TMEM91 | TMEM91 | 19756 | -0.711 | -0.0728 | No |
| 2266 | LRRC23 | LRRC23 | 19766 | -0.711 | -0.0728 | No |
| 2267 | DCLRE1A | DCLRE1A | 19799 | -0.714 | -0.0740 | No |
| 2268 | RAPGEF4 | RAPGEF4 | 19808 | -0.716 | -0.0739 | No |
| 2269 | RELL1 | RELL1 | 19837 | -0.732 | -0.0749 | No |
| 2270 | NAB2 | NAB2 | 19840 | -0.733 | -0.0746 | No |
| 2271 | CDC7 | CDC7 | 19881 | -0.754 | -0.0761 | No |
| 2272 | ADAMTS4 | ADAMTS4 | 19883 | -0.756 | -0.0757 | No |
| 2273 | ZCCHC3 | ZCCHC3 | 19886 | -0.759 | -0.0754 | No |
| 2274 | JUN | JUN | 19922 | -0.780 | -0.0767 | No |
| 2275 | NAB1 | NAB1 | 19947 | -0.795 | -0.0774 | No |
| 2276 | TRIM9 | TRIM9 | 19957 | -0.801 | -0.0774 | No |
| 2277 | DNAJB5 | DNAJB5 | 19978 | -0.812 | -0.0779 | No |
| 2278 | SCHIP1 | SCHIP1 | 19979 | -0.812 | -0.0774 | No |
| 2279 | ZBTB12 | ZBTB12 | 19993 | -0.820 | -0.0775 | No |
| 2280 | ENO2 | ENO2 | 20010 | -0.831 | -0.0778 | No |
| 2281 | SPRY1 | SPRY1 | 20015 | -0.835 | -0.0775 | No |
| 2282 | NEK4 | NEK4 | 20046 | -0.854 | -0.0785 | No |
| 2283 | GGCT | GGCT | 20047 | -0.854 | -0.0780 | No |
| 2284 | ALG10B | ALG10B | 20080 | -0.880 | -0.0791 | No |
| 2285 | MUC19 | MUC19 | 20086 | -0.881 | -0.0788 | No |
| 2286 | CIART | CIART | 20131 | -0.911 | -0.0805 | No |
| 2287 | RNY1 | RNY1 | 20149 | -0.920 | -0.0808 | No |
| 2288 | GSTA4 | GSTA4 | 20151 | -0.923 | -0.0803 | No |
| 2289 | FANCI | FANCI | 20161 | -0.925 | -0.0802 | No |
| 2290 | WFIKKN1 | WFIKKN1 | 20162 | -0.925 | -0.0796 | No |
| 2291 | CLYBL | CLYBL | 20168 | -0.931 | -0.0793 | No |
| 2292 | ASAH2 | ASAH2 | 20186 | -0.941 | -0.0796 | No |
| 2293 | SLC7A3 | SLC7A3 | 20188 | -0.941 | -0.0791 | No |
| 2294 | RTN4RL1 | RTN4RL1 | 20198 | -0.950 | -0.0789 | No |
| 2295 | GUCA1B | GUCA1B | 20199 | -0.950 | -0.0784 | No |
| 2296 | PGBD5 | PGBD5 | 20211 | -0.958 | -0.0783 | No |
| 2297 | KCNF1 | KCNF1 | 20222 | -0.960 | -0.0783 | No |
| 2298 | PEAK1 | PEAK1 | 20246 | -0.979 | -0.0788 | No |
| 2299 | POLD1 | POLD1 | 20248 | -0.980 | -0.0783 | No |
| 2300 | RND1 | RND1 | 20252 | -0.982 | -0.0778 | No |
| 2301 | TANGO6 | TANGO6 | 20255 | -0.984 | -0.0773 | No |
| 2302 | DISP2 | DISP2 | 20279 | -0.996 | -0.0779 | No |
| 2303 | ALKBH2 | ALKBH2 | 20290 | -1.004 | -0.0778 | No |
| 2304 | ING4 | ING4 | 20296 | -1.006 | -0.0774 | No |
| 2305 | MOB3B | MOB3B | 20315 | -1.022 | -0.0777 | No |
| 2306 | FUT7 | FUT7 | 20323 | -1.029 | -0.0774 | No |
| 2307 | CDKN1A | CDKN1A | 20334 | -1.035 | -0.0773 | No |
| 2308 | FAM217B | FAM217B | 20350 | -1.043 | -0.0774 | No |
| 2309 | CEBPA | CEBPA | 20375 | -1.064 | -0.0780 | No |
| 2310 | MLIP | MLIP | 20443 | -1.103 | -0.0807 | No |
| 2311 | BEND3 | BEND3 | 20458 | -1.113 | -0.0808 | No |
| 2312 | HSD11B2 | HSD11B2 | 20481 | -1.137 | -0.0812 | No |
| 2313 | MAP3K15 | MAP3K15 | 20485 | -1.139 | -0.0806 | No |
| 2314 | PSRC1 | PSRC1 | 20510 | -1.152 | -0.0812 | No |
| 2315 | GAL3ST3 | GAL3ST3 | 20564 | -1.198 | -0.0831 | No |
| 2316 | TLE6 | TLE6 | 20591 | -1.213 | -0.0837 | No |
| 2317 | TRNP1 | TRNP1 | 20593 | -1.214 | -0.0830 | No |
| 2318 | TGIF1 | TGIF1 | 20630 | -1.242 | -0.0841 | No |
| 2319 | NRG2 | NRG2 | 20684 | -1.299 | -0.0860 | No |
| 2320 | GAP43 | GAP43 | 20696 | -1.306 | -0.0857 | No |
| 2321 | RASA4 | RASA4 | 20697 | -1.306 | -0.0849 | No |
| 2322 | LRRTM3 | LRRTM3 | 20698 | -1.306 | -0.0841 | No |
| 2323 | SCML4 | SCML4 | 20703 | -1.316 | -0.0835 | No |
| 2324 | TLE2 | TLE2 | 20716 | -1.326 | -0.0833 | No |
| 2325 | POLE | POLE | 20723 | -1.330 | -0.0828 | No |
| 2326 | PAQR9 | PAQR9 | 20741 | -1.339 | -0.0829 | No |
| 2327 | PTPN5 | PTPN5 | 20756 | -1.351 | -0.0827 | No |
| 2328 | AS3MT | AS3MT | 20760 | -1.356 | -0.0821 | No |
| 2329 | CALML4 | CALML4 | 20766 | -1.361 | -0.0815 | No |
| 2330 | GLIPR2 | GLIPR2 | 20778 | -1.368 | -0.0812 | No |
| 2331 | MPPE1 | MPPE1 | 20792 | -1.381 | -0.0810 | No |
| 2332 | PDE4A | PDE4A | 20861 | -1.447 | -0.0836 | No |
| 2333 | ARC | ARC | 20863 | -1.448 | -0.0828 | No |
| 2334 | SEMA5A | SEMA5A | 20903 | -1.484 | -0.0838 | No |
| 2335 | XXYLT1 | XXYLT1 | 20919 | -1.505 | -0.0837 | No |
| 2336 | TSPAN15 | TSPAN15 | 20929 | -1.515 | -0.0832 | No |
| 2337 | SNORD59A | SNORD59A | 20938 | -1.528 | -0.0827 | No |
| 2338 | GNG4 | GNG4 | 20954 | -1.543 | -0.0825 | No |
| 2339 | ARHGAP20 | ARHGAP20 | 20959 | -1.547 | -0.0818 | No |
| 2340 | LGI2 | LGI2 | 20973 | -1.567 | -0.0815 | No |
| 2341 | CAMK1D | CAMK1D | 20974 | -1.568 | -0.0805 | No |
| 2342 | MAOB | MAOB | 20984 | -1.598 | -0.0800 | No |
| 2343 | NECAB1 | NECAB1 | 20993 | -1.613 | -0.0794 | No |
| 2344 | DOCK4 | DOCK4 | 21005 | -1.621 | -0.0790 | No |
| 2345 | NDRG1 | NDRG1 | 21010 | -1.628 | -0.0782 | No |
| 2346 | FIBIN | FIBIN | 21025 | -1.648 | -0.0779 | No |
| 2347 | SORCS3 | SORCS3 | 21038 | -1.662 | -0.0775 | No |
| 2348 | NRTN | NRTN | 21039 | -1.663 | -0.0765 | No |
| 2349 | DENND4C | DENND4C | 21040 | -1.665 | -0.0755 | No |
| 2350 | TDRP | TDRP | 21059 | -1.697 | -0.0754 | No |
| 2351 | GLB1L2 | GLB1L2 | 21070 | -1.716 | -0.0748 | No |
| 2352 | CAMK1G | CAMK1G | 21084 | -1.730 | -0.0744 | No |
| 2353 | MBNL3 | MBNL3 | 21086 | -1.735 | -0.0734 | No |
| 2354 | MATN2 | MATN2 | 21088 | -1.736 | -0.0724 | No |
| 2355 | E2F2 | E2F2 | 21090 | -1.742 | -0.0714 | No |
| 2356 | TRIM69 | TRIM69 | 21103 | -1.758 | -0.0709 | No |
| 2357 | ADGRG6 | ADGRG6 | 21125 | -1.769 | -0.0709 | No |
| 2358 | PDE1A | PDE1A | 21131 | -1.778 | -0.0701 | No |
| 2359 | OTX2 | OTX2 | 21134 | -1.788 | -0.0691 | No |
| 2360 | ST3GAL5 | ST3GAL5 | 21135 | -1.788 | -0.0680 | No |
| 2361 | ANGPTL2 | ANGPTL2 | 21151 | -1.819 | -0.0677 | No |
| 2362 | ZBTB16 | ZBTB16 | 21178 | -1.845 | -0.0679 | No |
| 2363 | ARFGEF3 | ARFGEF3 | 21185 | -1.855 | -0.0670 | No |
| 2364 | CHST8 | CHST8 | 21238 | -1.925 | -0.0685 | No |
| 2365 | ADGRA1 | ADGRA1 | 21250 | -1.956 | -0.0679 | No |
| 2366 | HAPLN4 | HAPLN4 | 21251 | -1.956 | -0.0667 | No |
| 2367 | P2RX3 | P2RX3 | 21254 | -1.957 | -0.0656 | No |
| 2368 | CTH | CTH | 21256 | -1.958 | -0.0644 | No |
| 2369 | ALDH1A2 | ALDH1A2 | 21309 | -2.024 | -0.0658 | No |
| 2370 | SYCE2 | SYCE2 | 21325 | -2.053 | -0.0654 | No |
| 2371 | KNTC1 | KNTC1 | 21331 | -2.067 | -0.0643 | No |
| 2372 | LRRK2 | LRRK2 | 21346 | -2.082 | -0.0638 | No |
| 2373 | SPRY4 | SPRY4 | 21353 | -2.094 | -0.0628 | No |
| 2374 | IL33 | IL33 | 21369 | -2.140 | -0.0623 | No |
| 2375 | NRN1 | NRN1 | 21370 | -2.141 | -0.0610 | No |
| 2376 | NLRC3 | NLRC3 | 21385 | -2.148 | -0.0604 | No |
| 2377 | DLG2 | DLG2 | 21387 | -2.148 | -0.0591 | No |
| 2378 | ADGRV1 | ADGRV1 | 21437 | -2.239 | -0.0602 | No |
| 2379 | ARHGAP6 | ARHGAP6 | 21489 | -2.377 | -0.0614 | No |
| 2380 | CBFA2T3 | CBFA2T3 | 21542 | -2.554 | -0.0624 | No |
| 2381 | AHRR | AHRR | 21558 | -2.592 | -0.0616 | No |
| 2382 | SNCB | SNCB | 21567 | -2.592 | -0.0604 | No |
| 2383 | ATP13A4 | ATP13A4 | 21620 | -2.638 | -0.0615 | No |
| 2384 | SMYD1 | SMYD1 | 21651 | -2.773 | -0.0613 | No |
| 2385 | LRRC10B | LRRC10B | 21657 | -2.773 | -0.0599 | No |
| 2386 | PLTP | PLTP | 21667 | -2.829 | -0.0586 | No |
| 2387 | PROM2 | PROM2 | 21677 | -2.868 | -0.0573 | No |
| 2388 | DPP6 | DPP6 | 21694 | -2.868 | -0.0564 | No |
| 2389 | MXD3 | MXD3 | 21733 | -2.991 | -0.0565 | No |
| 2390 | LEFTY1 | LEFTY1 | 21771 | -3.215 | -0.0564 | No |
| 2391 | CLEC2D | CLEC2D | 21801 | -3.527 | -0.0557 | No |
| 2392 | SYT3 | SYT3 | 21803 | -3.544 | -0.0536 | No |
| 2393 | ATG9B | ATG9B | 21834 | -4.077 | -0.0526 | No |
| 2394 | GPLD1 | GPLD1 | 21848 | -4.405 | -0.0506 | No |
| 2395 | LMX1B | LMX1B | 21853 | -4.405 | -0.0481 | No |
| 2396 | ATP13A5 | ATP13A5 | 21873 | -4.579 | -0.0463 | No |
| 2397 | SHISA6 | SHISA6 | 21874 | -4.616 | -0.0435 | No |
| 2398 | CGREF1 | CGREF1 | 21883 | -4.984 | -0.0409 | No |
| 2399 | HRASLS | HRASLS | 21892 | -5.000 | -0.0382 | No |
| 2400 | SLC40A1 | SLC40A1 | 21895 | -5.000 | -0.0353 | No |
| 2401 | BEAN1 | BEAN1 | 21902 | -5.000 | -0.0326 | No |
| 2402 | COLCA2 | COLCA2 | 21916 | -5.000 | -0.0302 | No |
| 2403 | VPREB3 | VPREB3 | 21930 | -5.000 | -0.0278 | No |
| 2404 | GRIN2D | GRIN2D | 21932 | -5.000 | -0.0248 | No |
| 2405 | TEX15 | TEX15 | 21940 | -5.000 | -0.0221 | No |
| 2406 | STAC2 | STAC2 | 21947 | -5.000 | -0.0194 | No |
| 2407 | CRHR1 | CRHR1 | 21949 | -5.000 | -0.0164 | No |
| 2408 | SLC16A10 | SLC16A10 | 21951 | -5.000 | -0.0134 | No |
| 2409 | MASP1 | MASP1 | 21964 | -5.000 | -0.0109 | No |
| 2410 | EMILIN2 | EMILIN2 | 21967 | -5.000 | -0.0080 | No |
| 2411 | CRISPLD1 | CRISPLD1 | 21971 | -5.000 | -0.0051 | No |
| 2412 | ADAM23 | ADAM23 | 21972 | -5.000 | -0.0021 | No |
| 2413 | GSTM4 | GSTM4 | 21982 | -5.000 | 0.0005 | No |
| 2414 | CREB3L4 | CREB3L4 | 21983 | -5.000 | 0.0036 | No |
| 2415 | KLHDC8A | KLHDC8A | 22017 | -5.000 | 0.0050 | No |
| 2416 | PRRT4 | PRRT4 | 22075 | -5.000 | 0.0051 | No |