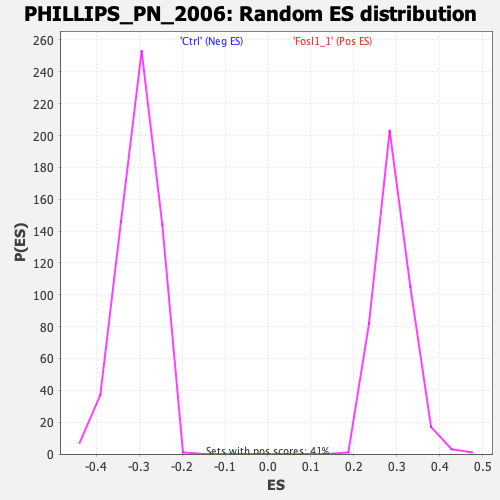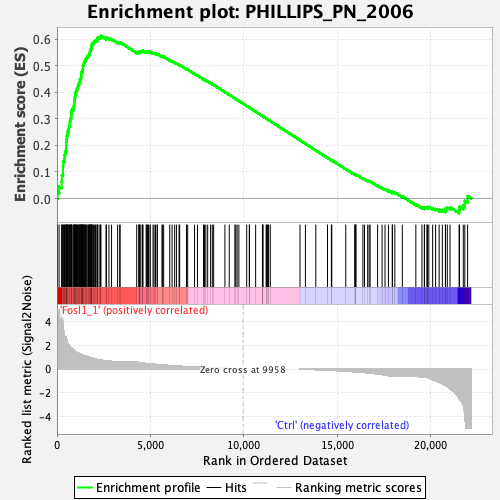
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Fosl1_1 |
| GeneSet | PHILLIPS_PN_2006 |
| Enrichment Score (ES) | 0.612356 |
| Normalized Enrichment Score (NES) | 2.1017513 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FAIM2 | FAIM2 | 27 | 5.000 | 0.0236 | Yes |
| 2 | DOK6 | DOK6 | 111 | 5.000 | 0.0447 | Yes |
| 3 | SCN3A | SCN3A | 248 | 5.000 | 0.0633 | Yes |
| 4 | FGF12 | FGF12 | 260 | 4.993 | 0.0876 | Yes |
| 5 | DSCAML1 | DSCAML1 | 320 | 3.766 | 0.1036 | Yes |
| 6 | HEY2 | HEY2 | 321 | 3.765 | 0.1223 | Yes |
| 7 | FXYD6 | FXYD6 | 329 | 3.517 | 0.1395 | Yes |
| 8 | NALCN | NALCN | 387 | 2.865 | 0.1511 | Yes |
| 9 | NOG | NOG | 405 | 2.756 | 0.1640 | Yes |
| 10 | PHACTR3 | PHACTR3 | 437 | 2.725 | 0.1761 | Yes |
| 11 | PRKCZ | PRKCZ | 486 | 2.653 | 0.1871 | Yes |
| 12 | SLC1A1 | SLC1A1 | 491 | 2.601 | 0.1998 | Yes |
| 13 | ANKS1B | ANKS1B | 495 | 2.588 | 0.2126 | Yes |
| 14 | SHD | SHD | 517 | 2.470 | 0.2239 | Yes |
| 15 | FSTL5 | FSTL5 | 527 | 2.410 | 0.2354 | Yes |
| 16 | ID4 | ID4 | 566 | 2.195 | 0.2446 | Yes |
| 17 | KIF5A | KIF5A | 581 | 2.140 | 0.2546 | Yes |
| 18 | EPHB1 | EPHB1 | 632 | 2.059 | 0.2625 | Yes |
| 19 | DSCAM | DSCAM | 640 | 2.035 | 0.2723 | Yes |
| 20 | IL17D | IL17D | 685 | 1.937 | 0.2799 | Yes |
| 21 | MAPK8IP2 | MAPK8IP2 | 691 | 1.924 | 0.2893 | Yes |
| 22 | PLEKHB1 | PLEKHB1 | 710 | 1.882 | 0.2978 | Yes |
| 23 | SCG3 | SCG3 | 752 | 1.812 | 0.3049 | Yes |
| 24 | GABBR2 | GABBR2 | 759 | 1.807 | 0.3136 | Yes |
| 25 | NAP1L3 | NAP1L3 | 771 | 1.788 | 0.3220 | Yes |
| 26 | GRIK4 | GRIK4 | 775 | 1.781 | 0.3307 | Yes |
| 27 | HLF | HLF | 816 | 1.709 | 0.3374 | Yes |
| 28 | SORBS1 | SORBS1 | 901 | 1.611 | 0.3416 | Yes |
| 29 | GAD1 | GAD1 | 905 | 1.603 | 0.3494 | Yes |
| 30 | DNM3 | DNM3 | 922 | 1.571 | 0.3565 | Yes |
| 31 | OMG | OMG | 923 | 1.571 | 0.3643 | Yes |
| 32 | RAB11FIP4 | RAB11FIP4 | 941 | 1.552 | 0.3712 | Yes |
| 33 | CSMD3 | CSMD3 | 948 | 1.544 | 0.3786 | Yes |
| 34 | DUSP26 | DUSP26 | 969 | 1.509 | 0.3852 | Yes |
| 35 | NKAIN4 | NKAIN4 | 977 | 1.498 | 0.3923 | Yes |
| 36 | JPH3 | JPH3 | 991 | 1.483 | 0.3991 | Yes |
| 37 | GPR158 | GPR158 | 1026 | 1.444 | 0.4047 | Yes |
| 38 | CMTM5 | CMTM5 | 1048 | 1.417 | 0.4108 | Yes |
| 39 | KCNN3 | KCNN3 | 1091 | 1.384 | 0.4157 | Yes |
| 40 | ATP6V1G2 | ATP6V1G2 | 1120 | 1.358 | 0.4212 | Yes |
| 41 | SOX6 | SOX6 | 1128 | 1.349 | 0.4276 | Yes |
| 42 | SNAP91 | SNAP91 | 1167 | 1.323 | 0.4324 | Yes |
| 43 | TMEM100 | TMEM100 | 1196 | 1.296 | 0.4376 | Yes |
| 44 | PCSK6 | PCSK6 | 1227 | 1.264 | 0.4425 | Yes |
| 45 | RGS9 | RGS9 | 1230 | 1.263 | 0.4487 | Yes |
| 46 | CNTN1 | CNTN1 | 1268 | 1.242 | 0.4532 | Yes |
| 47 | TTYH1 | TTYH1 | 1289 | 1.228 | 0.4584 | Yes |
| 48 | KCNB1 | KCNB1 | 1294 | 1.225 | 0.4643 | Yes |
| 49 | NTRK2 | NTRK2 | 1299 | 1.221 | 0.4701 | Yes |
| 50 | RPRM | RPRM | 1309 | 1.214 | 0.4758 | Yes |
| 51 | PHYHIPL | PHYHIPL | 1368 | 1.172 | 0.4790 | Yes |
| 52 | MN1 | MN1 | 1372 | 1.170 | 0.4846 | Yes |
| 53 | ELMO1 | ELMO1 | 1386 | 1.166 | 0.4898 | Yes |
| 54 | ENHO | ENHO | 1387 | 1.165 | 0.4956 | Yes |
| 55 | SEZ6L | SEZ6L | 1388 | 1.163 | 0.5014 | Yes |
| 56 | TAL1 | TAL1 | 1432 | 1.142 | 0.5051 | Yes |
| 57 | BCAN | BCAN | 1442 | 1.138 | 0.5103 | Yes |
| 58 | SH3GL2 | SH3GL2 | 1464 | 1.132 | 0.5150 | Yes |
| 59 | FBXO2 | FBXO2 | 1505 | 1.107 | 0.5187 | Yes |
| 60 | SLITRK2 | SLITRK2 | 1520 | 1.097 | 0.5235 | Yes |
| 61 | GRIA2 | GRIA2 | 1556 | 1.083 | 0.5273 | Yes |
| 62 | SLITRK5 | SLITRK5 | 1594 | 1.065 | 0.5309 | Yes |
| 63 | OPCML | OPCML | 1617 | 1.052 | 0.5351 | Yes |
| 64 | NRSN1 | NRSN1 | 1687 | 1.013 | 0.5370 | Yes |
| 65 | DLL3 | DLL3 | 1711 | 1.002 | 0.5409 | Yes |
| 66 | B3GAT1 | B3GAT1 | 1738 | 0.988 | 0.5446 | Yes |
| 67 | GABRB3 | GABRB3 | 1749 | 0.981 | 0.5491 | Yes |
| 68 | SOX8 | SOX8 | 1771 | 0.971 | 0.5529 | Yes |
| 69 | SMOC1 | SMOC1 | 1772 | 0.971 | 0.5577 | Yes |
| 70 | RTN1 | RTN1 | 1805 | 0.958 | 0.5610 | Yes |
| 71 | GRIA4 | GRIA4 | 1825 | 0.945 | 0.5649 | Yes |
| 72 | CSDC2 | CSDC2 | 1838 | 0.940 | 0.5690 | Yes |
| 73 | NDRG2 | NDRG2 | 1850 | 0.932 | 0.5731 | Yes |
| 74 | C1QL1 | C1QL1 | 1865 | 0.923 | 0.5771 | Yes |
| 75 | LGR5 | LGR5 | 1868 | 0.922 | 0.5816 | Yes |
| 76 | MAPT | MAPT | 1887 | 0.917 | 0.5853 | Yes |
| 77 | PSD | PSD | 1960 | 0.878 | 0.5864 | Yes |
| 78 | OLIG2 | OLIG2 | 1982 | 0.868 | 0.5897 | Yes |
| 79 | ALDOC | ALDOC | 2027 | 0.851 | 0.5920 | Yes |
| 80 | KCTD4 | KCTD4 | 2058 | 0.836 | 0.5947 | Yes |
| 81 | TIMP4 | TIMP4 | 2138 | 0.805 | 0.5951 | Yes |
| 82 | OLIG1 | OLIG1 | 2160 | 0.796 | 0.5981 | Yes |
| 83 | EHD3 | EHD3 | 2161 | 0.795 | 0.6021 | Yes |
| 84 | DLL1 | DLL1 | 2175 | 0.790 | 0.6054 | Yes |
| 85 | KIF1A | KIF1A | 2255 | 0.757 | 0.6056 | Yes |
| 86 | PDK2 | PDK2 | 2327 | 0.736 | 0.6060 | Yes |
| 87 | TMEM59L | TMEM59L | 2333 | 0.735 | 0.6094 | Yes |
| 88 | SMAD9 | SMAD9 | 2350 | 0.734 | 0.6124 | Yes |
| 89 | APOE | APOE | 2626 | 0.685 | 0.6032 | No |
| 90 | FAM13C | FAM13C | 2643 | 0.679 | 0.6059 | No |
| 91 | KSR2 | KSR2 | 2781 | 0.654 | 0.6029 | No |
| 92 | ASCL1 | ASCL1 | 2920 | 0.636 | 0.5998 | No |
| 93 | FUT9 | FUT9 | 3243 | 0.617 | 0.5882 | No |
| 94 | CNTN3 | CNTN3 | 3359 | 0.602 | 0.5860 | No |
| 95 | P2RY13 | P2RY13 | 3376 | 0.602 | 0.5882 | No |
| 96 | THRA | THRA | 4266 | 0.578 | 0.5506 | No |
| 97 | ASB13 | ASB13 | 4387 | 0.544 | 0.5479 | No |
| 98 | KIF21B | KIF21B | 4393 | 0.542 | 0.5503 | No |
| 99 | CRTC1 | CRTC1 | 4400 | 0.538 | 0.5527 | No |
| 100 | GRID1 | GRID1 | 4425 | 0.531 | 0.5543 | No |
| 101 | FRY | FRY | 4487 | 0.519 | 0.5541 | No |
| 102 | REPS2 | REPS2 | 4570 | 0.498 | 0.5528 | No |
| 103 | FBXL15 | FBXL15 | 4588 | 0.493 | 0.5545 | No |
| 104 | MMP16 | MMP16 | 4597 | 0.491 | 0.5566 | No |
| 105 | PCSK1N | PCSK1N | 4774 | 0.459 | 0.5509 | No |
| 106 | CALCRL | CALCRL | 4794 | 0.456 | 0.5523 | No |
| 107 | GABBR1 | GABBR1 | 4840 | 0.447 | 0.5524 | No |
| 108 | FAM19A5 | FAM19A5 | 4896 | 0.438 | 0.5521 | No |
| 109 | GNAL | GNAL | 4922 | 0.432 | 0.5531 | No |
| 110 | JPH4 | JPH4 | 4998 | 0.421 | 0.5518 | No |
| 111 | TMOD2 | TMOD2 | 5141 | 0.400 | 0.5473 | No |
| 112 | PTGDS | PTGDS | 5194 | 0.391 | 0.5469 | No |
| 113 | DNAJC12 | DNAJC12 | 5272 | 0.380 | 0.5453 | No |
| 114 | RUNDC3A | RUNDC3A | 5364 | 0.369 | 0.5430 | No |
| 115 | IKZF5 | IKZF5 | 5366 | 0.368 | 0.5448 | No |
| 116 | SNRPN | SNRPN | 5624 | 0.332 | 0.5347 | No |
| 117 | SGSM1 | SGSM1 | 5638 | 0.331 | 0.5358 | No |
| 118 | DTX4 | DTX4 | 5709 | 0.320 | 0.5342 | No |
| 119 | NCAM1 | NCAM1 | 6035 | 0.282 | 0.5208 | No |
| 120 | MARCH8 | MARCH8 | 6160 | 0.268 | 0.5165 | No |
| 121 | GNAO1 | GNAO1 | 6298 | 0.254 | 0.5115 | No |
| 122 | CYFIP2 | CYFIP2 | 6393 | 0.243 | 0.5084 | No |
| 123 | CBX7 | CBX7 | 6533 | 0.232 | 0.5033 | No |
| 124 | PKNOX2 | PKNOX2 | 6567 | 0.229 | 0.5029 | No |
| 125 | FSD1 | FSD1 | 6931 | 0.195 | 0.4873 | No |
| 126 | HIP1R | HIP1R | 6994 | 0.191 | 0.4855 | No |
| 127 | ALDH5A1 | ALDH5A1 | 7362 | 0.159 | 0.4696 | No |
| 128 | LRRC4 | LRRC4 | 7517 | 0.146 | 0.4633 | No |
| 129 | CCNK | CCNK | 7841 | 0.130 | 0.4492 | No |
| 130 | HDAC5 | HDAC5 | 7898 | 0.126 | 0.4473 | No |
| 131 | ARL3 | ARL3 | 7967 | 0.122 | 0.4448 | No |
| 132 | PARD3 | PARD3 | 8065 | 0.115 | 0.4410 | No |
| 133 | GLUD1 | GLUD1 | 8221 | 0.104 | 0.4344 | No |
| 134 | PID1 | PID1 | 8312 | 0.098 | 0.4308 | No |
| 135 | RPL37 | RPL37 | 8385 | 0.094 | 0.4280 | No |
| 136 | EPB41L2 | EPB41L2 | 8989 | 0.053 | 0.4008 | No |
| 137 | FAM110B | FAM110B | 9219 | 0.038 | 0.3906 | No |
| 138 | ZC3H12B | ZC3H12B | 9522 | 0.017 | 0.3770 | No |
| 139 | NUMA1 | NUMA1 | 9570 | 0.014 | 0.3749 | No |
| 140 | ARPP21 | ARPP21 | 9640 | 0.010 | 0.3718 | No |
| 141 | FGF13 | FGF13 | 9735 | 0.009 | 0.3676 | No |
| 142 | SEC31B | SEC31B | 10154 | 0.000 | 0.3485 | No |
| 143 | CALN1 | CALN1 | 10293 | 0.000 | 0.3423 | No |
| 144 | GALNT13 | GALNT13 | 10305 | 0.000 | 0.3418 | No |
| 145 | FAM155A | FAM155A | 10633 | 0.000 | 0.3269 | No |
| 146 | OVOL1 | OVOL1 | 10996 | 0.000 | 0.3104 | No |
| 147 | SLIT1 | SLIT1 | 11002 | 0.000 | 0.3102 | No |
| 148 | FGF14 | FGF14 | 11012 | 0.000 | 0.3098 | No |
| 149 | SPHKAP | SPHKAP | 11025 | 0.000 | 0.3092 | No |
| 150 | USH1C | USH1C | 11196 | 0.000 | 0.3015 | No |
| 151 | GABRA3 | GABRA3 | 11211 | 0.000 | 0.3009 | No |
| 152 | NEU4 | NEU4 | 11275 | 0.000 | 0.2980 | No |
| 153 | SSTR1 | SSTR1 | 11298 | 0.000 | 0.2970 | No |
| 154 | STOX1 | STOX1 | 11327 | 0.000 | 0.2957 | No |
| 155 | NRG3 | NRG3 | 11418 | 0.000 | 0.2916 | No |
| 156 | RPL13 | RPL13 | 13007 | -0.000 | 0.2194 | No |
| 157 | GPRC5B | GPRC5B | 13298 | -0.009 | 0.2062 | No |
| 158 | RAC3 | RAC3 | 13853 | -0.050 | 0.1813 | No |
| 159 | GAB2 | GAB2 | 14479 | -0.094 | 0.1533 | No |
| 160 | SLC1A4 | SLC1A4 | 14687 | -0.111 | 0.1444 | No |
| 161 | ZCCHC24 | ZCCHC24 | 14708 | -0.112 | 0.1441 | No |
| 162 | AP2B1 | AP2B1 | 15457 | -0.175 | 0.1109 | No |
| 163 | BEND7 | BEND7 | 15934 | -0.227 | 0.0904 | No |
| 164 | ABHD6 | ABHD6 | 15975 | -0.231 | 0.0897 | No |
| 165 | RIMS2 | RIMS2 | 15988 | -0.233 | 0.0903 | No |
| 166 | GGTA1 | GGTA1 | 16002 | -0.233 | 0.0909 | No |
| 167 | TNKS2 | TNKS2 | 16373 | -0.278 | 0.0754 | No |
| 168 | ABLIM1 | ABLIM1 | 16460 | -0.291 | 0.0730 | No |
| 169 | PDK4 | PDK4 | 16630 | -0.317 | 0.0668 | No |
| 170 | TPCN2 | TPCN2 | 16706 | -0.330 | 0.0651 | No |
| 171 | P2RX7 | P2RX7 | 16761 | -0.340 | 0.0643 | No |
| 172 | SGCG | SGCG | 17160 | -0.411 | 0.0482 | No |
| 173 | FLRT1 | FLRT1 | 17402 | -0.462 | 0.0396 | No |
| 174 | ATRNL1 | ATRNL1 | 17559 | -0.504 | 0.0350 | No |
| 175 | NET1 | NET1 | 17741 | -0.564 | 0.0295 | No |
| 176 | SLC25A21 | SLC25A21 | 17949 | -0.599 | 0.0231 | No |
| 177 | DPP10 | DPP10 | 17953 | -0.599 | 0.0259 | No |
| 178 | HS3ST4 | HS3ST4 | 18083 | -0.599 | 0.0230 | No |
| 179 | ABLIM3 | ABLIM3 | 18484 | -0.602 | 0.0078 | No |
| 180 | MAF | MAF | 19208 | -0.630 | -0.0219 | No |
| 181 | SUSD5 | SUSD5 | 19534 | -0.665 | -0.0334 | No |
| 182 | TPM1 | TPM1 | 19665 | -0.691 | -0.0359 | No |
| 183 | PLCB1 | PLCB1 | 19667 | -0.692 | -0.0325 | No |
| 184 | GFRA1 | GFRA1 | 19801 | -0.714 | -0.0350 | No |
| 185 | SSTR2 | SSTR2 | 19827 | -0.727 | -0.0326 | No |
| 186 | PKP4 | PKP4 | 19902 | -0.768 | -0.0321 | No |
| 187 | HSPA12A | HSPA12A | 20108 | -0.895 | -0.0370 | No |
| 188 | WNT7B | WNT7B | 20265 | -0.991 | -0.0392 | No |
| 189 | NTN4 | NTN4 | 20455 | -1.110 | -0.0423 | No |
| 190 | RASL10A | RASL10A | 20626 | -1.239 | -0.0438 | No |
| 191 | MPPE1 | MPPE1 | 20792 | -1.381 | -0.0445 | No |
| 192 | CRYAB | CRYAB | 20793 | -1.381 | -0.0376 | No |
| 193 | BMP2 | BMP2 | 20892 | -1.469 | -0.0348 | No |
| 194 | SORCS3 | SORCS3 | 21038 | -1.662 | -0.0331 | No |
| 195 | GRIA1 | GRIA1 | 21523 | -2.483 | -0.0428 | No |
| 196 | RASSF4 | RASSF4 | 21548 | -2.579 | -0.0311 | No |
| 197 | ARHGAP22 | ARHGAP22 | 21747 | -3.040 | -0.0250 | No |
| 198 | KCNQ5 | KCNQ5 | 21829 | -3.952 | -0.0091 | No |
| 199 | F2 | F2 | 21978 | -5.000 | 0.0090 | No |