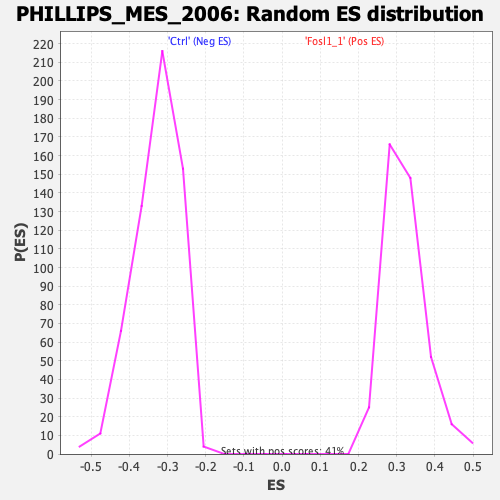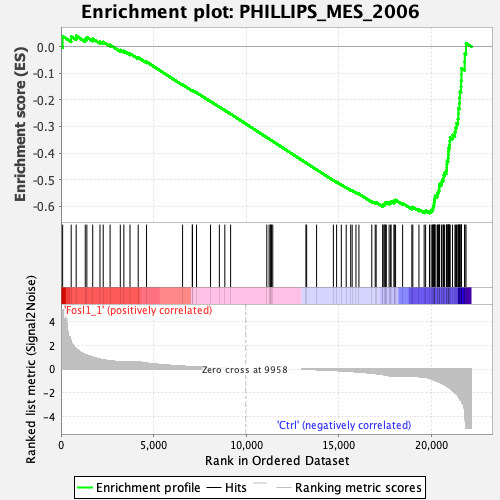
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Ctrl |
| GeneSet | PHILLIPS_MES_2006 |
| Enrichment Score (ES) | -0.6263039 |
| Normalized Enrichment Score (NES) | -1.91194 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RYR3 | RYR3 | 89 | 5.000 | 0.0394 | No |
| 2 | ANGPTL4 | ANGPTL4 | 550 | 2.260 | 0.0382 | No |
| 3 | VWA1 | VWA1 | 820 | 1.703 | 0.0408 | No |
| 4 | LRRC29 | LRRC29 | 1310 | 1.212 | 0.0292 | No |
| 5 | NRP2 | NRP2 | 1397 | 1.160 | 0.0354 | No |
| 6 | BACE2 | BACE2 | 1714 | 0.998 | 0.0298 | No |
| 7 | CECR2 | CECR2 | 2106 | 0.818 | 0.0192 | No |
| 8 | SHROOM3 | SHROOM3 | 2281 | 0.748 | 0.0178 | No |
| 9 | MYL9 | MYL9 | 2652 | 0.677 | 0.0069 | No |
| 10 | EPAS1 | EPAS1 | 3203 | 0.630 | -0.0125 | No |
| 11 | PLA2G5 | PLA2G5 | 3396 | 0.602 | -0.0160 | No |
| 12 | SLC22A18 | SLC22A18 | 3728 | 0.602 | -0.0258 | No |
| 13 | GGN | GGN | 4171 | 0.601 | -0.0406 | No |
| 14 | PLEKHF1 | PLEKHF1 | 4621 | 0.485 | -0.0567 | No |
| 15 | OSMR | OSMR | 6568 | 0.229 | -0.1429 | No |
| 16 | HOMER3 | HOMER3 | 7091 | 0.181 | -0.1650 | No |
| 17 | TTC38 | TTC38 | 7105 | 0.181 | -0.1640 | No |
| 18 | FAM20C | FAM20C | 7322 | 0.162 | -0.1724 | No |
| 19 | PAPPA | PAPPA | 8079 | 0.114 | -0.2057 | No |
| 20 | SLC12A9 | SLC12A9 | 8555 | 0.083 | -0.2265 | No |
| 21 | TWF1 | TWF1 | 8850 | 0.063 | -0.2392 | No |
| 22 | TRABD | TRABD | 9163 | 0.042 | -0.2530 | No |
| 23 | SPOCD1 | SPOCD1 | 11118 | 0.000 | -0.3415 | No |
| 24 | TAGLN | TAGLN | 11242 | 0.000 | -0.3471 | No |
| 25 | ACTA2 | ACTA2 | 11306 | 0.000 | -0.3500 | No |
| 26 | ICAM1 | ICAM1 | 11338 | 0.000 | -0.3514 | No |
| 27 | ANPEP | ANPEP | 11375 | 0.000 | -0.3530 | No |
| 28 | HSD3B7 | HSD3B7 | 11441 | 0.000 | -0.3560 | No |
| 29 | LZTS1 | LZTS1 | 13232 | -0.005 | -0.4370 | No |
| 30 | FGFRL1 | FGFRL1 | 13271 | -0.007 | -0.4387 | No |
| 31 | ECE1 | ECE1 | 13808 | -0.047 | -0.4626 | No |
| 32 | RRAS | RRAS | 14715 | -0.113 | -0.5026 | No |
| 33 | UNC93B1 | UNC93B1 | 14889 | -0.125 | -0.5094 | No |
| 34 | TPP1 | TPP1 | 15144 | -0.144 | -0.5196 | No |
| 35 | BHLHE40 | BHLHE40 | 15408 | -0.170 | -0.5301 | No |
| 36 | RAB34 | RAB34 | 15651 | -0.195 | -0.5394 | No |
| 37 | NRP1 | NRP1 | 15730 | -0.203 | -0.5411 | No |
| 38 | DEF6 | DEF6 | 15935 | -0.227 | -0.5484 | No |
| 39 | FLNA | FLNA | 16095 | -0.243 | -0.5535 | No |
| 40 | TRIM56 | TRIM56 | 16790 | -0.346 | -0.5819 | No |
| 41 | CD151 | CD151 | 16975 | -0.377 | -0.5870 | No |
| 42 | NCLN | NCLN | 17026 | -0.386 | -0.5859 | No |
| 43 | ZYX | ZYX | 17383 | -0.458 | -0.5981 | No |
| 44 | RRBP1 | RRBP1 | 17390 | -0.459 | -0.5943 | No |
| 45 | PDPN | PDPN | 17456 | -0.474 | -0.5932 | No |
| 46 | EFEMP2 | EFEMP2 | 17509 | -0.488 | -0.5913 | No |
| 47 | KLF16 | KLF16 | 17519 | -0.494 | -0.5874 | No |
| 48 | SOCS3 | SOCS3 | 17575 | -0.509 | -0.5855 | No |
| 49 | MAP2K3 | MAP2K3 | 17727 | -0.559 | -0.5874 | No |
| 50 | NEURL2 | NEURL2 | 17813 | -0.588 | -0.5862 | No |
| 51 | RHOJ | RHOJ | 17828 | -0.595 | -0.5816 | No |
| 52 | FPR2 | FPR2 | 17994 | -0.599 | -0.5839 | No |
| 53 | SLC39A8 | SLC39A8 | 18002 | -0.599 | -0.5790 | No |
| 54 | THBD | THBD | 18075 | -0.599 | -0.5771 | No |
| 55 | SALL4 | SALL4 | 18456 | -0.602 | -0.5891 | No |
| 56 | IFITM2 | IFITM2 | 18954 | -0.603 | -0.6063 | No |
| 57 | HRH1 | HRH1 | 19007 | -0.619 | -0.6033 | No |
| 58 | PDLIM7 | PDLIM7 | 19338 | -0.640 | -0.6127 | No |
| 59 | PDGFA | PDGFA | 19619 | -0.685 | -0.6194 | No |
| 60 | PMEPA1 | PMEPA1 | 19698 | -0.705 | -0.6168 | No |
| 61 | SGSH | SGSH | 19908 | -0.771 | -0.6196 | Yes |
| 62 | TNC | TNC | 19997 | -0.820 | -0.6165 | Yes |
| 63 | MYH9 | MYH9 | 20066 | -0.867 | -0.6120 | Yes |
| 64 | MVP | MVP | 20100 | -0.891 | -0.6057 | Yes |
| 65 | ITGA7 | ITGA7 | 20111 | -0.896 | -0.5984 | Yes |
| 66 | FOSL2 | FOSL2 | 20154 | -0.923 | -0.5923 | Yes |
| 67 | TRIM47 | TRIM47 | 20157 | -0.923 | -0.5844 | Yes |
| 68 | SLC25A37 | SLC25A37 | 20181 | -0.938 | -0.5772 | Yes |
| 69 | COL4A1 | COL4A1 | 20197 | -0.950 | -0.5697 | Yes |
| 70 | FBN1 | FBN1 | 20207 | -0.956 | -0.5618 | Yes |
| 71 | SERPINH1 | SERPINH1 | 20318 | -1.025 | -0.5578 | Yes |
| 72 | ACTN1 | ACTN1 | 20344 | -1.040 | -0.5499 | Yes |
| 73 | PDLIM4 | PDLIM4 | 20391 | -1.075 | -0.5427 | Yes |
| 74 | SHC1 | SHC1 | 20429 | -1.093 | -0.5348 | Yes |
| 75 | DLC1 | DLC1 | 20431 | -1.095 | -0.5254 | Yes |
| 76 | CD248 | CD248 | 20456 | -1.111 | -0.5168 | Yes |
| 77 | JUNB | JUNB | 20559 | -1.193 | -0.5110 | Yes |
| 78 | FES | FES | 20586 | -1.212 | -0.5017 | Yes |
| 79 | IFITM3 | IFITM3 | 20666 | -1.284 | -0.4941 | Yes |
| 80 | EMP1 | EMP1 | 20675 | -1.290 | -0.4832 | Yes |
| 81 | COL4A2 | COL4A2 | 20724 | -1.331 | -0.4739 | Yes |
| 82 | PDGFRL | PDGFRL | 20822 | -1.421 | -0.4659 | Yes |
| 83 | BCL3 | BCL3 | 20841 | -1.433 | -0.4543 | Yes |
| 84 | OSBPL3 | OSBPL3 | 20845 | -1.434 | -0.4419 | Yes |
| 85 | STEAP3 | STEAP3 | 20857 | -1.442 | -0.4299 | Yes |
| 86 | FLT1 | FLT1 | 20913 | -1.497 | -0.4194 | Yes |
| 87 | LPAR1 | LPAR1 | 20921 | -1.506 | -0.4066 | Yes |
| 88 | EFNB2 | EFNB2 | 20931 | -1.518 | -0.3938 | Yes |
| 89 | METTL7B | METTL7B | 20941 | -1.531 | -0.3809 | Yes |
| 90 | ITGA5 | ITGA5 | 20995 | -1.615 | -0.3693 | Yes |
| 91 | LIF | LIF | 21001 | -1.620 | -0.3554 | Yes |
| 92 | TIMP1 | TIMP1 | 21019 | -1.640 | -0.3419 | Yes |
| 93 | C1QTNF1 | C1QTNF1 | 21152 | -1.819 | -0.3321 | Yes |
| 94 | MMP14 | MMP14 | 21282 | -1.998 | -0.3206 | Yes |
| 95 | EHD2 | EHD2 | 21320 | -2.040 | -0.3045 | Yes |
| 96 | LRRC32 | LRRC32 | 21368 | -2.140 | -0.2880 | Yes |
| 97 | SERPINE1 | SERPINE1 | 21444 | -2.262 | -0.2718 | Yes |
| 98 | ITGA3 | ITGA3 | 21463 | -2.316 | -0.2525 | Yes |
| 99 | EMP3 | EMP3 | 21469 | -2.325 | -0.2325 | Yes |
| 100 | S100A11 | S100A11 | 21526 | -2.486 | -0.2134 | Yes |
| 101 | ANGPT2 | ANGPT2 | 21536 | -2.525 | -0.1919 | Yes |
| 102 | RUNX1 | RUNX1 | 21549 | -2.581 | -0.1700 | Yes |
| 103 | PARP10 | PARP10 | 21611 | -2.599 | -0.1501 | Yes |
| 104 | SLC16A3 | SLC16A3 | 21618 | -2.631 | -0.1275 | Yes |
| 105 | C1RL | C1RL | 21634 | -2.707 | -0.1047 | Yes |
| 106 | SBNO2 | SBNO2 | 21636 | -2.708 | -0.0812 | Yes |
| 107 | PLAU | PLAU | 21807 | -3.644 | -0.0572 | Yes |
| 108 | ESM1 | ESM1 | 21815 | -3.692 | -0.0254 | Yes |
| 109 | PLAUR | PLAUR | 21880 | -4.804 | 0.0134 | Yes |