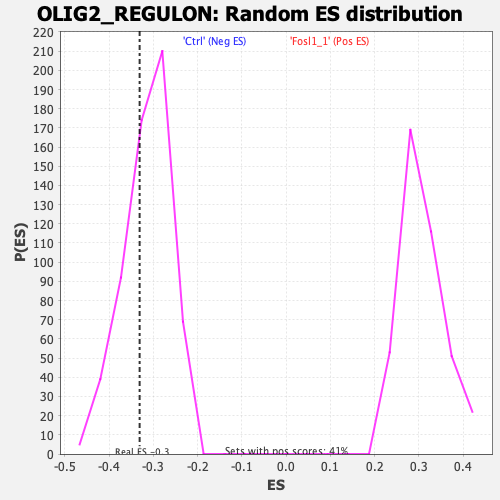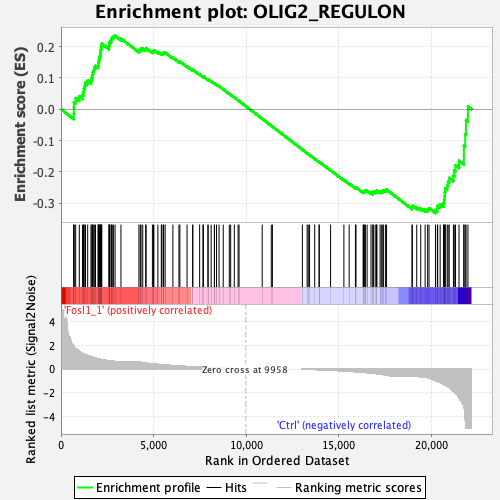
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Ctrl |
| GeneSet | OLIG2_REGULON |
| Enrichment Score (ES) | -0.33128822 |
| Normalized Enrichment Score (NES) | -1.052342 |
| Nominal p-value | 0.34295416 |
| FDR q-value | 0.39326277 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RLBP1 | RLBP1 | 692 | 1.924 | -0.0131 | No |
| 2 | NLGN3 | NLGN3 | 696 | 1.919 | 0.0050 | No |
| 3 | PNMA2 | PNMA2 | 704 | 1.902 | 0.0227 | No |
| 4 | PODXL2 | PODXL2 | 793 | 1.759 | 0.0355 | No |
| 5 | BCHE | BCHE | 988 | 1.489 | 0.0408 | No |
| 6 | ST3GAL6 | ST3GAL6 | 1162 | 1.328 | 0.0456 | No |
| 7 | ABAT | ABAT | 1213 | 1.274 | 0.0554 | No |
| 8 | GPR19 | GPR19 | 1234 | 1.258 | 0.0665 | No |
| 9 | TTYH1 | TTYH1 | 1289 | 1.228 | 0.0757 | No |
| 10 | FAM131B | FAM131B | 1325 | 1.204 | 0.0855 | No |
| 11 | BCAN | BCAN | 1442 | 1.138 | 0.0911 | No |
| 12 | TMPRSS5 | TMPRSS5 | 1619 | 1.052 | 0.0931 | No |
| 13 | VGLL4 | VGLL4 | 1666 | 1.027 | 0.1008 | No |
| 14 | PTPRZ1 | PTPRZ1 | 1701 | 1.008 | 0.1088 | No |
| 15 | MAP2 | MAP2 | 1712 | 1.001 | 0.1179 | No |
| 16 | RASSF2 | RASSF2 | 1773 | 0.970 | 0.1243 | No |
| 17 | TUBB2B | TUBB2B | 1795 | 0.961 | 0.1325 | No |
| 18 | PPFIBP2 | PPFIBP2 | 1864 | 0.924 | 0.1382 | No |
| 19 | GAB1 | GAB1 | 2006 | 0.859 | 0.1400 | No |
| 20 | S100B | S100B | 2014 | 0.856 | 0.1478 | No |
| 21 | PLLP | PLLP | 2039 | 0.845 | 0.1548 | No |
| 22 | FRMPD1 | FRMPD1 | 2060 | 0.834 | 0.1618 | No |
| 23 | CCDC28B | CCDC28B | 2083 | 0.827 | 0.1686 | No |
| 24 | SLC38A3 | SLC38A3 | 2128 | 0.809 | 0.1743 | No |
| 25 | TIMP4 | TIMP4 | 2138 | 0.805 | 0.1816 | No |
| 26 | BCAR3 | BCAR3 | 2141 | 0.804 | 0.1891 | No |
| 27 | CSPG5 | CSPG5 | 2171 | 0.792 | 0.1953 | No |
| 28 | MARCKSL1 | MARCKSL1 | 2177 | 0.790 | 0.2026 | No |
| 29 | TRAF4 | TRAF4 | 2204 | 0.776 | 0.2088 | No |
| 30 | NOVA1 | NOVA1 | 2593 | 0.694 | 0.1978 | No |
| 31 | CITED1 | CITED1 | 2597 | 0.692 | 0.2042 | No |
| 32 | CHST11 | CHST11 | 2627 | 0.685 | 0.2094 | No |
| 33 | TBKBP1 | TBKBP1 | 2630 | 0.682 | 0.2158 | No |
| 34 | SLC4A4 | SLC4A4 | 2714 | 0.670 | 0.2184 | No |
| 35 | SOX11 | SOX11 | 2717 | 0.668 | 0.2247 | No |
| 36 | RBBP9 | RBBP9 | 2768 | 0.656 | 0.2286 | No |
| 37 | FEZ1 | FEZ1 | 2830 | 0.647 | 0.2320 | No |
| 38 | ASCL1 | ASCL1 | 2920 | 0.636 | 0.2340 | No |
| 39 | GNG7 | GNG7 | 3240 | 0.618 | 0.2254 | No |
| 40 | TUBA1A | TUBA1A | 4210 | 0.592 | 0.1871 | No |
| 41 | MAP3K1 | MAP3K1 | 4285 | 0.571 | 0.1891 | No |
| 42 | RGS20 | RGS20 | 4323 | 0.561 | 0.1928 | No |
| 43 | RAP1GAP | RAP1GAP | 4415 | 0.535 | 0.1937 | No |
| 44 | PICK1 | PICK1 | 4576 | 0.497 | 0.1912 | No |
| 45 | MMP16 | MMP16 | 4597 | 0.491 | 0.1950 | No |
| 46 | LMF1 | LMF1 | 4938 | 0.431 | 0.1836 | No |
| 47 | RASL12 | RASL12 | 4995 | 0.422 | 0.1851 | No |
| 48 | POLR2F | POLR2F | 5017 | 0.419 | 0.1881 | No |
| 49 | BMP2K | BMP2K | 5235 | 0.387 | 0.1820 | No |
| 50 | GPD1L | GPD1L | 5419 | 0.361 | 0.1771 | No |
| 51 | SYT11 | SYT11 | 5449 | 0.357 | 0.1792 | No |
| 52 | TRIB2 | TRIB2 | 5529 | 0.344 | 0.1789 | No |
| 53 | TMEM206 | TMEM206 | 5545 | 0.343 | 0.1814 | No |
| 54 | ATP6V0E2 | ATP6V0E2 | 5627 | 0.332 | 0.1809 | No |
| 55 | PHLPP1 | PHLPP1 | 6044 | 0.281 | 0.1647 | No |
| 56 | DBI | DBI | 6371 | 0.245 | 0.1522 | No |
| 57 | GPR68 | GPR68 | 6423 | 0.241 | 0.1522 | No |
| 58 | NME4 | NME4 | 6810 | 0.205 | 0.1366 | No |
| 59 | TBC1D22B | TBC1D22B | 7116 | 0.180 | 0.1245 | No |
| 60 | RAF1 | RAF1 | 7118 | 0.180 | 0.1262 | No |
| 61 | SNX27 | SNX27 | 7490 | 0.149 | 0.1108 | No |
| 62 | STARD7 | STARD7 | 7668 | 0.139 | 0.1041 | No |
| 63 | TMEM121 | TMEM121 | 7693 | 0.138 | 0.1043 | No |
| 64 | TRIM24 | TRIM24 | 7942 | 0.123 | 0.0942 | No |
| 65 | ENDOG | ENDOG | 7945 | 0.123 | 0.0953 | No |
| 66 | CYP1B1 | CYP1B1 | 8111 | 0.111 | 0.0888 | No |
| 67 | CSPG4 | CSPG4 | 8286 | 0.099 | 0.0819 | No |
| 68 | IDH1 | IDH1 | 8292 | 0.099 | 0.0826 | No |
| 69 | RBM38 | RBM38 | 8402 | 0.092 | 0.0785 | No |
| 70 | COL7A1 | COL7A1 | 8538 | 0.084 | 0.0732 | No |
| 71 | PIK3R2 | PIK3R2 | 8764 | 0.069 | 0.0637 | No |
| 72 | KANK1 | KANK1 | 9092 | 0.047 | 0.0493 | No |
| 73 | IRS1 | IRS1 | 9166 | 0.042 | 0.0464 | No |
| 74 | FSCN1 | FSCN1 | 9363 | 0.028 | 0.0377 | No |
| 75 | MAP2K5 | MAP2K5 | 9567 | 0.014 | 0.0287 | No |
| 76 | IFFO1 | IFFO1 | 9619 | 0.011 | 0.0264 | No |
| 77 | SRGN | SRGN | 10870 | 0.000 | -0.0303 | No |
| 78 | SLC19A3 | SLC19A3 | 11360 | 0.000 | -0.0525 | No |
| 79 | NINJ2 | NINJ2 | 11421 | 0.000 | -0.0552 | No |
| 80 | RAB27A | RAB27A | 13040 | -0.001 | -0.1286 | No |
| 81 | GPRC5B | GPRC5B | 13298 | -0.009 | -0.1402 | No |
| 82 | AKAP12 | AKAP12 | 13357 | -0.014 | -0.1427 | No |
| 83 | NFAT5 | NFAT5 | 13428 | -0.019 | -0.1457 | No |
| 84 | ATXN10 | ATXN10 | 13706 | -0.039 | -0.1579 | No |
| 85 | AGPAT5 | AGPAT5 | 13944 | -0.055 | -0.1681 | No |
| 86 | GNAZ | GNAZ | 13951 | -0.056 | -0.1678 | No |
| 87 | NDE1 | NDE1 | 14564 | -0.101 | -0.1946 | No |
| 88 | ELL2 | ELL2 | 15283 | -0.157 | -0.2257 | No |
| 89 | LAPTM4B | LAPTM4B | 15567 | -0.187 | -0.2368 | No |
| 90 | HOMER1 | HOMER1 | 15906 | -0.224 | -0.2500 | No |
| 91 | SYT17 | SYT17 | 15937 | -0.228 | -0.2492 | No |
| 92 | HIP1 | HIP1 | 16324 | -0.271 | -0.2641 | No |
| 93 | QDPR | QDPR | 16338 | -0.274 | -0.2621 | No |
| 94 | GLDC | GLDC | 16388 | -0.281 | -0.2617 | No |
| 95 | GOLM1 | GOLM1 | 16433 | -0.286 | -0.2610 | No |
| 96 | SPATA6 | SPATA6 | 16435 | -0.287 | -0.2583 | No |
| 97 | ATP9B | ATP9B | 16538 | -0.302 | -0.2600 | No |
| 98 | FZD7 | FZD7 | 16748 | -0.338 | -0.2663 | No |
| 99 | NES | NES | 16849 | -0.355 | -0.2675 | No |
| 100 | SHC3 | SHC3 | 16855 | -0.356 | -0.2643 | No |
| 101 | VASH1 | VASH1 | 16902 | -0.364 | -0.2629 | No |
| 102 | APBB2 | APBB2 | 17007 | -0.382 | -0.2640 | No |
| 103 | RRAS2 | RRAS2 | 17047 | -0.390 | -0.2621 | No |
| 104 | CCND2 | CCND2 | 17068 | -0.395 | -0.2592 | No |
| 105 | MFAP2 | MFAP2 | 17237 | -0.425 | -0.2628 | No |
| 106 | GAS1 | GAS1 | 17305 | -0.440 | -0.2617 | No |
| 107 | ZYX | ZYX | 17383 | -0.458 | -0.2608 | No |
| 108 | ETV5 | ETV5 | 17415 | -0.464 | -0.2578 | No |
| 109 | IRF1 | IRF1 | 17525 | -0.496 | -0.2581 | No |
| 110 | LEPROTL1 | LEPROTL1 | 17588 | -0.513 | -0.2560 | No |
| 111 | CREB5 | CREB5 | 18961 | -0.605 | -0.3125 | No |
| 112 | HES1 | HES1 | 18981 | -0.610 | -0.3076 | No |
| 113 | BAALC | BAALC | 19219 | -0.632 | -0.3123 | No |
| 114 | ART3 | ART3 | 19435 | -0.648 | -0.3159 | No |
| 115 | DNAH9 | DNAH9 | 19672 | -0.693 | -0.3200 | No |
| 116 | RAPGEF4 | RAPGEF4 | 19808 | -0.716 | -0.3194 | No |
| 117 | FSTL1 | FSTL1 | 19882 | -0.754 | -0.3155 | No |
| 118 | DUSP6 | DUSP6 | 20231 | -0.966 | -0.3221 | Yes |
| 119 | COL4A6 | COL4A6 | 20333 | -1.034 | -0.3169 | Yes |
| 120 | PHACTR2 | PHACTR2 | 20351 | -1.043 | -0.3077 | Yes |
| 121 | CD44 | CD44 | 20484 | -1.138 | -0.3029 | Yes |
| 122 | GCNT1 | GCNT1 | 20662 | -1.275 | -0.2988 | Yes |
| 123 | SLIT2 | SLIT2 | 20700 | -1.310 | -0.2880 | Yes |
| 124 | MYO1C | MYO1C | 20732 | -1.333 | -0.2768 | Yes |
| 125 | FAM114A1 | FAM114A1 | 20738 | -1.339 | -0.2643 | Yes |
| 126 | SLC10A3 | SLC10A3 | 20759 | -1.355 | -0.2523 | Yes |
| 127 | ARC | ARC | 20863 | -1.448 | -0.2432 | Yes |
| 128 | SDC2 | SDC2 | 20912 | -1.495 | -0.2312 | Yes |
| 129 | LGI2 | LGI2 | 20973 | -1.567 | -0.2190 | Yes |
| 130 | MYOF | MYOF | 21202 | -1.877 | -0.2115 | Yes |
| 131 | PTGS2 | PTGS2 | 21243 | -1.932 | -0.1950 | Yes |
| 132 | PTGER4 | PTGER4 | 21314 | -2.033 | -0.1789 | Yes |
| 133 | PRDM8 | PRDM8 | 21503 | -2.429 | -0.1643 | Yes |
| 134 | CAST | CAST | 21765 | -3.156 | -0.1462 | Yes |
| 135 | P4HA2 | P4HA2 | 21767 | -3.184 | -0.1159 | Yes |
| 136 | VEGFC | VEGFC | 21837 | -4.155 | -0.0796 | Yes |
| 137 | CGREF1 | CGREF1 | 21883 | -4.984 | -0.0343 | Yes |
| 138 | BNC2 | BNC2 | 21985 | -5.000 | 0.0087 | Yes |