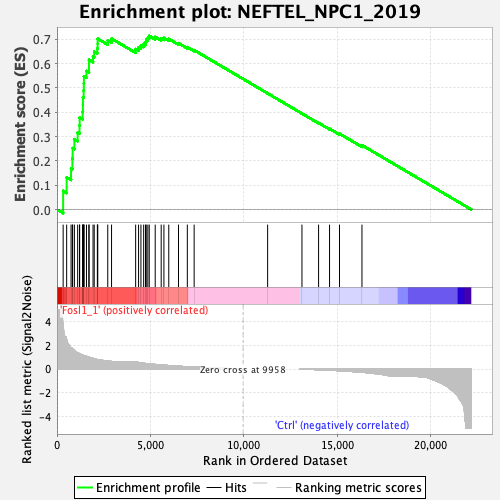
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Fosl1_1 |
| GeneSet | NEFTEL_NPC1_2019 |
| Enrichment Score (ES) | 0.7136113 |
| Normalized Enrichment Score (NES) | 1.9863435 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FXYD6 | FXYD6 | 329 | 3.517 | 0.0764 | Yes |
| 2 | SHD | SHD | 517 | 2.470 | 0.1320 | Yes |
| 3 | SCG3 | SCG3 | 752 | 1.812 | 0.1685 | Yes |
| 4 | ELAVL4 | ELAVL4 | 823 | 1.701 | 0.2095 | Yes |
| 5 | TNR | TNR | 837 | 1.676 | 0.2524 | Yes |
| 6 | TAGLN3 | TAGLN3 | 930 | 1.562 | 0.2887 | Yes |
| 7 | GRIK2 | GRIK2 | 1106 | 1.367 | 0.3163 | Yes |
| 8 | NXPH1 | NXPH1 | 1205 | 1.286 | 0.3452 | Yes |
| 9 | ABAT | ABAT | 1213 | 1.274 | 0.3780 | Yes |
| 10 | LRRN1 | LRRN1 | 1373 | 1.170 | 0.4012 | Yes |
| 11 | ELMO1 | ELMO1 | 1386 | 1.166 | 0.4309 | Yes |
| 12 | SEZ6L | SEZ6L | 1388 | 1.163 | 0.4610 | Yes |
| 13 | MYT1 | MYT1 | 1426 | 1.144 | 0.4890 | Yes |
| 14 | TUBB3 | TUBB3 | 1436 | 1.141 | 0.5182 | Yes |
| 15 | BCAN | BCAN | 1442 | 1.138 | 0.5475 | Yes |
| 16 | DCX | DCX | 1578 | 1.074 | 0.5693 | Yes |
| 17 | DLL3 | DLL3 | 1711 | 1.002 | 0.5893 | Yes |
| 18 | MAP2 | MAP2 | 1712 | 1.001 | 0.6153 | Yes |
| 19 | BTG2 | BTG2 | 1929 | 0.896 | 0.6288 | Yes |
| 20 | AMOTL2 | AMOTL2 | 1992 | 0.864 | 0.6484 | Yes |
| 21 | OLIG1 | OLIG1 | 2160 | 0.796 | 0.6615 | Yes |
| 22 | DLL1 | DLL1 | 2175 | 0.790 | 0.6814 | Yes |
| 23 | MARCKSL1 | MARCKSL1 | 2177 | 0.790 | 0.7019 | Yes |
| 24 | SOX11 | SOX11 | 2717 | 0.668 | 0.6949 | Yes |
| 25 | ASCL1 | ASCL1 | 2920 | 0.636 | 0.7022 | Yes |
| 26 | TUBA1A | TUBA1A | 4210 | 0.592 | 0.6594 | Yes |
| 27 | DBN1 | DBN1 | 4366 | 0.550 | 0.6666 | Yes |
| 28 | SEZ6 | SEZ6 | 4493 | 0.518 | 0.6744 | Yes |
| 29 | MLLT11 | MLLT11 | 4639 | 0.480 | 0.6803 | Yes |
| 30 | SERINC5 | SERINC5 | 4738 | 0.465 | 0.6879 | Yes |
| 31 | CHD7 | CHD7 | 4776 | 0.458 | 0.6982 | Yes |
| 32 | PFN2 | PFN2 | 4863 | 0.443 | 0.7058 | Yes |
| 33 | LMF1 | LMF1 | 4938 | 0.431 | 0.7136 | Yes |
| 34 | TSPAN13 | TSPAN13 | 5257 | 0.383 | 0.7092 | No |
| 35 | SOX4 | SOX4 | 5577 | 0.338 | 0.7035 | No |
| 36 | PTPRS | PTPRS | 5723 | 0.318 | 0.7052 | No |
| 37 | HES6 | HES6 | 5983 | 0.288 | 0.7010 | No |
| 38 | MEST | MEST | 6504 | 0.235 | 0.6836 | No |
| 39 | PCBP4 | PCBP4 | 6976 | 0.193 | 0.6674 | No |
| 40 | BEX1 | BEX1 | 7341 | 0.160 | 0.6551 | No |
| 41 | NEU4 | NEU4 | 11275 | 0.000 | 0.4773 | No |
| 42 | STMN1 | STMN1 | 13112 | -0.002 | 0.3944 | No |
| 43 | TCF12 | TCF12 | 14002 | -0.060 | 0.3558 | No |
| 44 | ETV1 | ETV1 | 14589 | -0.103 | 0.3320 | No |
| 45 | GLCCI1 | GLCCI1 | 15122 | -0.142 | 0.3117 | No |
| 46 | HIP1 | HIP1 | 16324 | -0.271 | 0.2644 | No |