Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | ranaseq_carol_ns_sel_exprs.ranaseq_carol_ns_sel_phenotype.cls #Fosl1_1_versus_Ctrl |
| Phenotype | ranaseq_carol_ns_sel_phenotype.cls#Fosl1_1_versus_Ctrl |
| Upregulated in class | Fosl1_1 |
| GeneSet | ASCL1_REGULON |
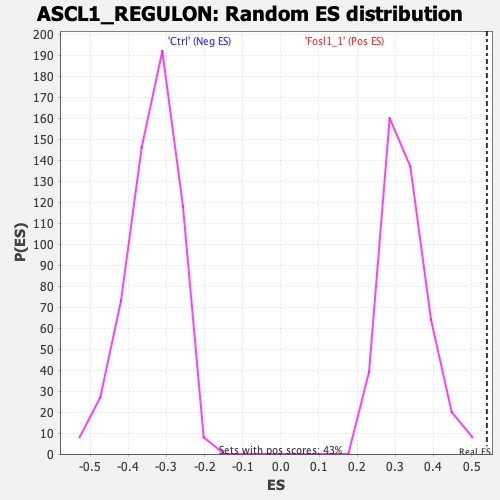
| Enrichment Score (ES) | 0.54014206 |
| Normalized Enrichment Score (NES) | 1.6614885 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0043267403 |
| FWER p-Value | 0.027 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PCSK2 | PCSK2 | 37 | 5.000 | 0.0537 | Yes |
| 2 | RAB33A | RAB33A | 369 | 2.975 | 0.0717 | Yes |
| 3 | RGR | RGR | 394 | 2.756 | 0.1012 | Yes |
| 4 | KCNJ10 | KCNJ10 | 518 | 2.470 | 0.1230 | Yes |
| 5 | ID4 | ID4 | 566 | 2.195 | 0.1452 | Yes |
| 6 | NLGN3 | NLGN3 | 696 | 1.919 | 0.1606 | Yes |
| 7 | SLC6A1 | SLC6A1 | 701 | 1.910 | 0.1816 | Yes |
| 8 | SCG3 | SCG3 | 752 | 1.812 | 0.1994 | Yes |
| 9 | GRIK4 | GRIK4 | 775 | 1.781 | 0.2181 | Yes |
| 10 | ELAVL4 | ELAVL4 | 823 | 1.701 | 0.2348 | Yes |
| 11 | GAD1 | GAD1 | 905 | 1.603 | 0.2489 | Yes |
| 12 | CRB1 | CRB1 | 992 | 1.482 | 0.2615 | Yes |
| 13 | ABAT | ABAT | 1213 | 1.274 | 0.2656 | Yes |
| 14 | DOCK10 | DOCK10 | 1225 | 1.266 | 0.2791 | Yes |
| 15 | TTYH1 | TTYH1 | 1289 | 1.228 | 0.2899 | Yes |
| 16 | ASTN1 | ASTN1 | 1356 | 1.178 | 0.3000 | Yes |
| 17 | LSAMP | LSAMP | 1410 | 1.153 | 0.3103 | Yes |
| 18 | MYT1 | MYT1 | 1426 | 1.144 | 0.3223 | Yes |
| 19 | BCAN | BCAN | 1442 | 1.138 | 0.3343 | Yes |
| 20 | GRIA2 | GRIA2 | 1556 | 1.083 | 0.3412 | Yes |
| 21 | DCX | DCX | 1578 | 1.074 | 0.3521 | Yes |
| 22 | SALL2 | SALL2 | 1644 | 1.036 | 0.3606 | Yes |
| 23 | GPR37L1 | GPR37L1 | 1665 | 1.027 | 0.3711 | Yes |
| 24 | CADM4 | CADM4 | 1692 | 1.012 | 0.3812 | Yes |
| 25 | MAP2 | MAP2 | 1712 | 1.001 | 0.3914 | Yes |
| 26 | B3GAT1 | B3GAT1 | 1738 | 0.988 | 0.4012 | Yes |
| 27 | SCRG1 | SCRG1 | 1740 | 0.986 | 0.4121 | Yes |
| 28 | TUBB2B | TUBB2B | 1795 | 0.961 | 0.4203 | Yes |
| 29 | NDRG2 | NDRG2 | 1850 | 0.932 | 0.4282 | Yes |
| 30 | ACSBG1 | ACSBG1 | 1879 | 0.919 | 0.4371 | Yes |
| 31 | NTRK3 | NTRK3 | 1905 | 0.908 | 0.4460 | Yes |
| 32 | MLC1 | MLC1 | 1922 | 0.899 | 0.4552 | Yes |
| 33 | BTG2 | BTG2 | 1929 | 0.896 | 0.4649 | Yes |
| 34 | OLIG2 | OLIG2 | 1982 | 0.868 | 0.4722 | Yes |
| 35 | KLF15 | KLF15 | 1988 | 0.865 | 0.4815 | Yes |
| 36 | BMP7 | BMP7 | 2038 | 0.845 | 0.4887 | Yes |
| 37 | NCAN | NCAN | 2117 | 0.813 | 0.4942 | Yes |
| 38 | ARHGEF9 | ARHGEF9 | 2122 | 0.810 | 0.5030 | Yes |
| 39 | CSPG5 | CSPG5 | 2171 | 0.792 | 0.5096 | Yes |
| 40 | JAKMIP2 | JAKMIP2 | 2186 | 0.788 | 0.5177 | Yes |
| 41 | PPP1R3C | PPP1R3C | 2203 | 0.778 | 0.5256 | Yes |
| 42 | REV3L | REV3L | 2219 | 0.769 | 0.5334 | Yes |
| 43 | CLASP2 | CLASP2 | 2418 | 0.727 | 0.5325 | Yes |
| 44 | PLXNB1 | PLXNB1 | 2573 | 0.699 | 0.5333 | Yes |
| 45 | CSGALNACT1 | CSGALNACT1 | 2651 | 0.677 | 0.5373 | Yes |
| 46 | PTN | PTN | 2750 | 0.659 | 0.5401 | Yes |
| 47 | LPAR4 | LPAR4 | 3249 | 0.615 | 0.5244 | No |
| 48 | HAPLN1 | HAPLN1 | 3331 | 0.602 | 0.5274 | No |
| 49 | MAP3K1 | MAP3K1 | 4285 | 0.571 | 0.4906 | No |
| 50 | CRMP1 | CRMP1 | 4419 | 0.534 | 0.4905 | No |
| 51 | SATB2 | SATB2 | 4820 | 0.451 | 0.4774 | No |
| 52 | MTSS1 | MTSS1 | 5291 | 0.378 | 0.4603 | No |
| 53 | PHLPP1 | PHLPP1 | 6044 | 0.281 | 0.4293 | No |
| 54 | GPM6B | GPM6B | 6372 | 0.245 | 0.4172 | No |
| 55 | PTP4A3 | PTP4A3 | 6629 | 0.222 | 0.4081 | No |
| 56 | PTK7 | PTK7 | 7455 | 0.151 | 0.3724 | No |
| 57 | SMARCD3 | SMARCD3 | 7748 | 0.134 | 0.3607 | No |
| 58 | PID1 | PID1 | 8312 | 0.098 | 0.3363 | No |
| 59 | SEPT2 | SEPT2 | 8582 | 0.081 | 0.3250 | No |
| 60 | RERE | RERE | 8855 | 0.063 | 0.3134 | No |
| 61 | BBS7 | BBS7 | 9035 | 0.050 | 0.3058 | No |
| 62 | WBP4 | WBP4 | 9303 | 0.032 | 0.2941 | No |
| 63 | TPBG | TPBG | 9585 | 0.013 | 0.2815 | No |
| 64 | SNTG1 | SNTG1 | 13131 | -0.002 | 0.1210 | No |
| 65 | MTMR12 | MTMR12 | 13444 | -0.020 | 0.1071 | No |
| 66 | CDK6 | CDK6 | 13463 | -0.021 | 0.1065 | No |
| 67 | UBN1 | UBN1 | 13604 | -0.031 | 0.1005 | No |
| 68 | LMO1 | LMO1 | 13915 | -0.054 | 0.0871 | No |
| 69 | TCF12 | TCF12 | 14002 | -0.060 | 0.0839 | No |
| 70 | SEPT7 | SEPT7 | 14260 | -0.079 | 0.0731 | No |
| 71 | RHOQ | RHOQ | 14433 | -0.090 | 0.0663 | No |
| 72 | GAB2 | GAB2 | 14479 | -0.094 | 0.0653 | No |
| 73 | ETV1 | ETV1 | 14589 | -0.103 | 0.0615 | No |
| 74 | ZCCHC24 | ZCCHC24 | 14708 | -0.112 | 0.0574 | No |
| 75 | TBC1D9 | TBC1D9 | 15921 | -0.226 | 0.0051 | No |
| 76 | GANAB | GANAB | 16219 | -0.258 | -0.0055 | No |
| 77 | MKNK1 | MKNK1 | 16336 | -0.273 | -0.0078 | No |
| 78 | P2RX7 | P2RX7 | 16761 | -0.340 | -0.0232 | No |
| 79 | BAG2 | BAG2 | 16801 | -0.348 | -0.0211 | No |
| 80 | PAK1 | PAK1 | 17195 | -0.417 | -0.0343 | No |
| 81 | OGFRL1 | OGFRL1 | 17550 | -0.502 | -0.0447 | No |
| 82 | RPE65 | RPE65 | 17904 | -0.599 | -0.0541 | No |
| 83 | PLS1 | PLS1 | 18400 | -0.601 | -0.0698 | No |
| 84 | ATP2B2 | ATP2B2 | 19742 | -0.709 | -0.1227 | No |
| 85 | TRIM9 | TRIM9 | 19957 | -0.801 | -0.1235 | No |
| 86 | ELF4 | ELF4 | 20859 | -1.446 | -0.1483 | No |
| 87 | IER3 | IER3 | 20879 | -1.462 | -0.1330 | No |
| 88 | SEMA5A | SEMA5A | 20903 | -1.484 | -0.1176 | No |
| 89 | MAOB | MAOB | 20984 | -1.598 | -0.1035 | No |
| 90 | MET | MET | 21431 | -2.217 | -0.0991 | No |
| 91 | COBL | COBL | 21546 | -2.571 | -0.0758 | No |
| 92 | RHBDF2 | RHBDF2 | 21846 | -4.405 | -0.0405 | No |
| 93 | MASP1 | MASP1 | 21964 | -5.000 | 0.0096 | No |