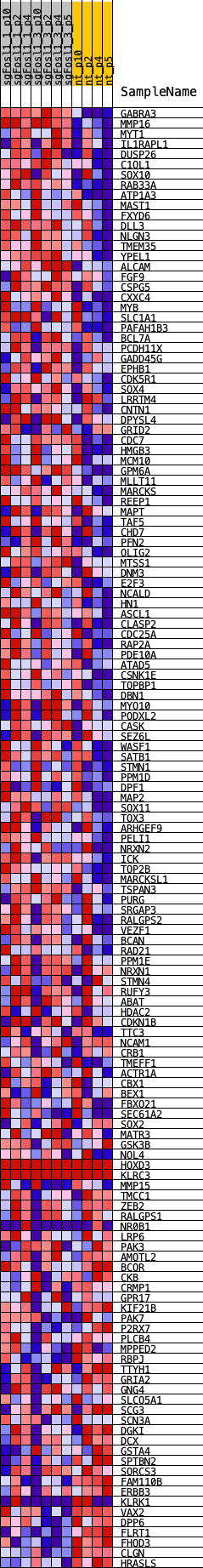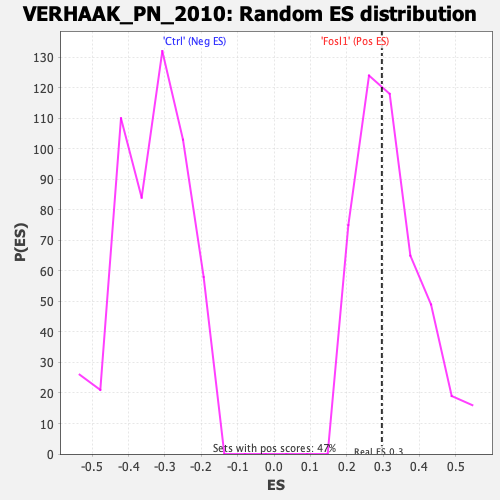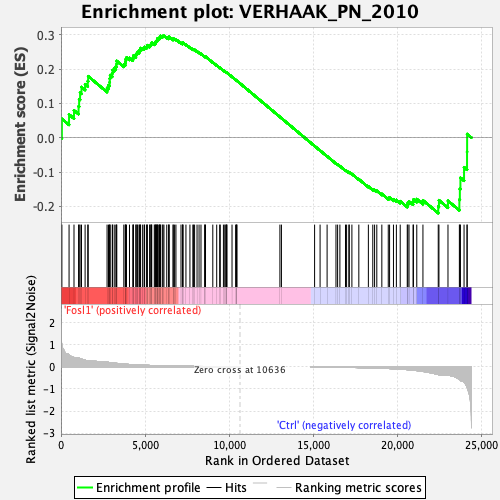
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Fosl1 |
| GeneSet | VERHAAK_PN_2010 |
| Enrichment Score (ES) | 0.29664814 |
| Normalized Enrichment Score (NES) | 0.9228229 |
| Nominal p-value | 0.55364805 |
| FDR q-value | 0.86363435 |
| FWER p-Value | 0.991 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GABRA3 | GABRA3 | 52 | 1.006 | 0.0548 | Yes |
| 2 | MMP16 | MMP16 | 479 | 0.536 | 0.0676 | Yes |
| 3 | MYT1 | MYT1 | 772 | 0.427 | 0.0797 | Yes |
| 4 | IL1RAPL1 | IL1RAPL1 | 1044 | 0.403 | 0.0913 | Yes |
| 5 | DUSP26 | DUSP26 | 1078 | 0.393 | 0.1122 | Yes |
| 6 | C1QL1 | C1QL1 | 1134 | 0.374 | 0.1311 | Yes |
| 7 | SOX10 | SOX10 | 1218 | 0.350 | 0.1476 | Yes |
| 8 | RAB33A | RAB33A | 1432 | 0.299 | 0.1557 | Yes |
| 9 | ATP1A3 | ATP1A3 | 1591 | 0.275 | 0.1647 | Yes |
| 10 | MAST1 | MAST1 | 1615 | 0.269 | 0.1790 | Yes |
| 11 | FXYD6 | FXYD6 | 2737 | 0.216 | 0.1450 | Yes |
| 12 | DLL3 | DLL3 | 2822 | 0.204 | 0.1531 | Yes |
| 13 | NLGN3 | NLGN3 | 2890 | 0.197 | 0.1615 | Yes |
| 14 | TMEM35 | TMEM35 | 2896 | 0.196 | 0.1724 | Yes |
| 15 | YPEL1 | YPEL1 | 2919 | 0.193 | 0.1824 | Yes |
| 16 | ALCAM | ALCAM | 3045 | 0.181 | 0.1875 | Yes |
| 17 | FGF9 | FGF9 | 3062 | 0.179 | 0.1970 | Yes |
| 18 | CSPG5 | CSPG5 | 3162 | 0.170 | 0.2025 | Yes |
| 19 | CXXC4 | CXXC4 | 3258 | 0.162 | 0.2077 | Yes |
| 20 | MYB | MYB | 3304 | 0.159 | 0.2149 | Yes |
| 21 | SLC1A1 | SLC1A1 | 3310 | 0.158 | 0.2236 | Yes |
| 22 | PAFAH1B3 | PAFAH1B3 | 3732 | 0.126 | 0.2134 | Yes |
| 23 | BCL7A | BCL7A | 3816 | 0.120 | 0.2167 | Yes |
| 24 | PCDH11X | PCDH11X | 3837 | 0.119 | 0.2226 | Yes |
| 25 | GADD45G | GADD45G | 3846 | 0.118 | 0.2290 | Yes |
| 26 | EPHB1 | EPHB1 | 3890 | 0.116 | 0.2338 | Yes |
| 27 | CDK5R1 | CDK5R1 | 4069 | 0.108 | 0.2325 | Yes |
| 28 | SOX4 | SOX4 | 4269 | 0.100 | 0.2300 | Yes |
| 29 | LRRTM4 | LRRTM4 | 4292 | 0.099 | 0.2347 | Yes |
| 30 | CNTN1 | CNTN1 | 4306 | 0.098 | 0.2397 | Yes |
| 31 | DPYSL4 | DPYSL4 | 4449 | 0.091 | 0.2389 | Yes |
| 32 | GRID2 | GRID2 | 4458 | 0.090 | 0.2437 | Yes |
| 33 | CDC7 | CDC7 | 4516 | 0.088 | 0.2463 | Yes |
| 34 | HMGB3 | HMGB3 | 4552 | 0.086 | 0.2498 | Yes |
| 35 | MCM10 | MCM10 | 4642 | 0.083 | 0.2508 | Yes |
| 36 | GPM6A | GPM6A | 4657 | 0.082 | 0.2549 | Yes |
| 37 | MLLT11 | MLLT11 | 4717 | 0.081 | 0.2570 | Yes |
| 38 | MARCKS | MARCKS | 4729 | 0.080 | 0.2611 | Yes |
| 39 | REEP1 | REEP1 | 4851 | 0.076 | 0.2604 | Yes |
| 40 | MAPT | MAPT | 4940 | 0.074 | 0.2610 | Yes |
| 41 | TAF5 | TAF5 | 4963 | 0.074 | 0.2642 | Yes |
| 42 | CHD7 | CHD7 | 5075 | 0.070 | 0.2636 | Yes |
| 43 | PFN2 | PFN2 | 5122 | 0.069 | 0.2657 | Yes |
| 44 | OLIG2 | OLIG2 | 5125 | 0.069 | 0.2695 | Yes |
| 45 | MTSS1 | MTSS1 | 5265 | 0.065 | 0.2674 | Yes |
| 46 | DNM3 | DNM3 | 5295 | 0.065 | 0.2699 | Yes |
| 47 | E2F3 | E2F3 | 5346 | 0.063 | 0.2714 | Yes |
| 48 | NCALD | NCALD | 5373 | 0.063 | 0.2739 | Yes |
| 49 | HN1 | HN1 | 5391 | 0.062 | 0.2767 | Yes |
| 50 | ASCL1 | ASCL1 | 5543 | 0.059 | 0.2738 | Yes |
| 51 | CLASP2 | CLASP2 | 5584 | 0.058 | 0.2755 | Yes |
| 52 | CDC25A | CDC25A | 5622 | 0.057 | 0.2772 | Yes |
| 53 | RAP2A | RAP2A | 5627 | 0.057 | 0.2803 | Yes |
| 54 | PDE10A | PDE10A | 5673 | 0.057 | 0.2816 | Yes |
| 55 | ATAD5 | ATAD5 | 5713 | 0.056 | 0.2832 | Yes |
| 56 | CSNK1E | CSNK1E | 5716 | 0.056 | 0.2863 | Yes |
| 57 | TOPBP1 | TOPBP1 | 5725 | 0.056 | 0.2891 | Yes |
| 58 | DBN1 | DBN1 | 5806 | 0.054 | 0.2888 | Yes |
| 59 | MYO10 | MYO10 | 5852 | 0.053 | 0.2900 | Yes |
| 60 | PODXL2 | PODXL2 | 5863 | 0.053 | 0.2926 | Yes |
| 61 | CASK | CASK | 5887 | 0.052 | 0.2946 | Yes |
| 62 | SEZ6L | SEZ6L | 5910 | 0.052 | 0.2966 | Yes |
| 63 | WASF1 | WASF1 | 6016 | 0.050 | 0.2951 | Yes |
| 64 | SATB1 | SATB1 | 6056 | 0.050 | 0.2963 | Yes |
| 65 | STMN1 | STMN1 | 6116 | 0.049 | 0.2966 | Yes |
| 66 | PPM1D | PPM1D | 6283 | 0.046 | 0.2924 | No |
| 67 | DPF1 | DPF1 | 6396 | 0.044 | 0.2903 | No |
| 68 | MAP2 | MAP2 | 6412 | 0.044 | 0.2921 | No |
| 69 | SOX11 | SOX11 | 6421 | 0.044 | 0.2943 | No |
| 70 | TOX3 | TOX3 | 6660 | 0.040 | 0.2867 | No |
| 71 | ARHGEF9 | ARHGEF9 | 6667 | 0.040 | 0.2887 | No |
| 72 | PELI1 | PELI1 | 6749 | 0.039 | 0.2876 | No |
| 73 | NRXN2 | NRXN2 | 6828 | 0.038 | 0.2865 | No |
| 74 | ICK | ICK | 7130 | 0.034 | 0.2760 | No |
| 75 | TOP2B | TOP2B | 7220 | 0.033 | 0.2742 | No |
| 76 | MARCKSL1 | MARCKSL1 | 7231 | 0.033 | 0.2757 | No |
| 77 | TSPAN3 | TSPAN3 | 7252 | 0.033 | 0.2767 | No |
| 78 | PURG | PURG | 7423 | 0.030 | 0.2714 | No |
| 79 | SRGAP3 | SRGAP3 | 7660 | 0.028 | 0.2632 | No |
| 80 | RALGPS2 | RALGPS2 | 7834 | 0.026 | 0.2575 | No |
| 81 | VEZF1 | VEZF1 | 7877 | 0.025 | 0.2572 | No |
| 82 | BCAN | BCAN | 7937 | 0.025 | 0.2562 | No |
| 83 | RAD21 | RAD21 | 8083 | 0.023 | 0.2515 | No |
| 84 | PPM1E | PPM1E | 8202 | 0.022 | 0.2479 | No |
| 85 | NRXN1 | NRXN1 | 8317 | 0.020 | 0.2443 | No |
| 86 | STMN4 | STMN4 | 8543 | 0.018 | 0.2361 | No |
| 87 | RUFY3 | RUFY3 | 8545 | 0.018 | 0.2371 | No |
| 88 | ABAT | ABAT | 8586 | 0.018 | 0.2364 | No |
| 89 | HDAC2 | HDAC2 | 9019 | 0.014 | 0.2194 | No |
| 90 | CDKN1B | CDKN1B | 9251 | 0.012 | 0.2106 | No |
| 91 | TTC3 | TTC3 | 9429 | 0.010 | 0.2039 | No |
| 92 | NCAM1 | NCAM1 | 9461 | 0.010 | 0.2032 | No |
| 93 | CRB1 | CRB1 | 9650 | 0.008 | 0.1959 | No |
| 94 | TMEFF1 | TMEFF1 | 9738 | 0.008 | 0.1927 | No |
| 95 | ACTR1A | ACTR1A | 9756 | 0.007 | 0.1924 | No |
| 96 | CBX1 | CBX1 | 9841 | 0.007 | 0.1894 | No |
| 97 | BEX1 | BEX1 | 9845 | 0.007 | 0.1896 | No |
| 98 | FBXO21 | FBXO21 | 9851 | 0.007 | 0.1898 | No |
| 99 | SEC61A2 | SEC61A2 | 10161 | 0.004 | 0.1773 | No |
| 100 | SOX2 | SOX2 | 10374 | 0.002 | 0.1687 | No |
| 101 | MATR3 | MATR3 | 10434 | 0.002 | 0.1663 | No |
| 102 | GSK3B | GSK3B | 10437 | 0.002 | 0.1663 | No |
| 103 | NOL4 | NOL4 | 10448 | 0.002 | 0.1660 | No |
| 104 | HOXD3 | HOXD3 | 13006 | 0.000 | 0.0605 | No |
| 105 | KLRC3 | KLRC3 | 13097 | 0.000 | 0.0568 | No |
| 106 | MMP15 | MMP15 | 15067 | -0.002 | -0.0243 | No |
| 107 | TMCC1 | TMCC1 | 15393 | -0.005 | -0.0374 | No |
| 108 | ZEB2 | ZEB2 | 15810 | -0.009 | -0.0541 | No |
| 109 | RALGPS1 | RALGPS1 | 16337 | -0.014 | -0.0750 | No |
| 110 | NR0B1 | NR0B1 | 16434 | -0.015 | -0.0781 | No |
| 111 | LRP6 | LRP6 | 16570 | -0.016 | -0.0827 | No |
| 112 | PAK3 | PAK3 | 16903 | -0.020 | -0.0953 | No |
| 113 | AMOTL2 | AMOTL2 | 16968 | -0.021 | -0.0968 | No |
| 114 | BCOR | BCOR | 17104 | -0.022 | -0.1011 | No |
| 115 | CKB | CKB | 17128 | -0.023 | -0.1007 | No |
| 116 | CRMP1 | CRMP1 | 17283 | -0.025 | -0.1057 | No |
| 117 | GPR17 | GPR17 | 17695 | -0.030 | -0.1209 | No |
| 118 | KIF21B | KIF21B | 18262 | -0.039 | -0.1421 | No |
| 119 | PAK7 | PAK7 | 18524 | -0.044 | -0.1503 | No |
| 120 | P2RX7 | P2RX7 | 18631 | -0.046 | -0.1521 | No |
| 121 | PLCB4 | PLCB4 | 18763 | -0.049 | -0.1547 | No |
| 122 | MPPED2 | MPPED2 | 19062 | -0.057 | -0.1637 | No |
| 123 | RBPJ | RBPJ | 19444 | -0.070 | -0.1755 | No |
| 124 | TTYH1 | TTYH1 | 19522 | -0.072 | -0.1746 | No |
| 125 | GRIA2 | GRIA2 | 19753 | -0.082 | -0.1794 | No |
| 126 | GNG4 | GNG4 | 19926 | -0.090 | -0.1814 | No |
| 127 | SLCO5A1 | SLCO5A1 | 20154 | -0.101 | -0.1851 | No |
| 128 | SCG3 | SCG3 | 20572 | -0.121 | -0.1954 | No |
| 129 | SCN3A | SCN3A | 20606 | -0.123 | -0.1898 | No |
| 130 | DGKI | DGKI | 20677 | -0.127 | -0.1856 | No |
| 131 | DCX | DCX | 20918 | -0.142 | -0.1874 | No |
| 132 | GSTA4 | GSTA4 | 20940 | -0.145 | -0.1800 | No |
| 133 | SPTBN2 | SPTBN2 | 21138 | -0.164 | -0.1789 | No |
| 134 | SORCS3 | SORCS3 | 21504 | -0.204 | -0.1824 | No |
| 135 | FAM110B | FAM110B | 22413 | -0.344 | -0.2004 | No |
| 136 | ERBB3 | ERBB3 | 22455 | -0.351 | -0.1822 | No |
| 137 | KLRK1 | KLRK1 | 22992 | -0.361 | -0.1839 | No |
| 138 | VAX2 | VAX2 | 23669 | -0.554 | -0.1804 | No |
| 139 | DPP6 | DPP6 | 23698 | -0.572 | -0.1492 | No |
| 140 | FLRT1 | FLRT1 | 23731 | -0.597 | -0.1167 | No |
| 141 | FHOD3 | FHOD3 | 23947 | -0.695 | -0.0862 | No |
| 142 | CLGN | CLGN | 24125 | -0.920 | -0.0414 | No |
| 143 | HRASLS | HRASLS | 24130 | -0.922 | 0.0106 | No |