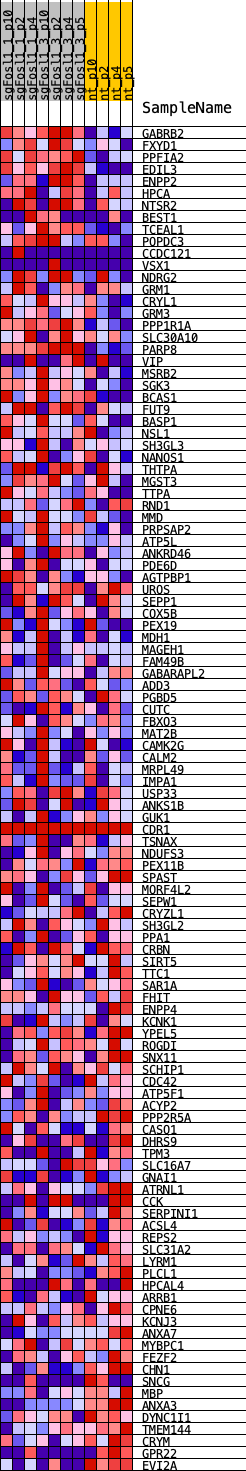
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | VERHAAK_NEU_2010 |
| Enrichment Score (ES) | -0.3042703 |
| Normalized Enrichment Score (NES) | -0.94737345 |
| Nominal p-value | 0.54285717 |
| FDR q-value | 0.6436056 |
| FWER p-Value | 0.991 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GABRB2 | GABRB2 | 212 | 0.676 | 0.0353 | No |
| 2 | FXYD1 | FXYD1 | 234 | 0.651 | 0.0768 | No |
| 3 | PPFIA2 | PPFIA2 | 483 | 0.533 | 0.1013 | No |
| 4 | EDIL3 | EDIL3 | 1115 | 0.382 | 0.1002 | No |
| 5 | ENPP2 | ENPP2 | 1225 | 0.348 | 0.1184 | No |
| 6 | HPCA | HPCA | 1422 | 0.301 | 0.1299 | No |
| 7 | NTSR2 | NTSR2 | 1590 | 0.275 | 0.1409 | No |
| 8 | BEST1 | BEST1 | 1633 | 0.266 | 0.1565 | No |
| 9 | TCEAL1 | TCEAL1 | 1641 | 0.265 | 0.1735 | No |
| 10 | POPDC3 | POPDC3 | 1753 | 0.255 | 0.1855 | No |
| 11 | CCDC121 | CCDC121 | 1933 | 0.250 | 0.1944 | No |
| 12 | VSX1 | VSX1 | 2570 | 0.240 | 0.1839 | No |
| 13 | NDRG2 | NDRG2 | 2656 | 0.228 | 0.1952 | No |
| 14 | GRM1 | GRM1 | 2696 | 0.222 | 0.2081 | No |
| 15 | CRYL1 | CRYL1 | 2775 | 0.210 | 0.2186 | No |
| 16 | GRM3 | GRM3 | 3003 | 0.185 | 0.2213 | No |
| 17 | PPP1R1A | PPP1R1A | 3018 | 0.183 | 0.2327 | No |
| 18 | SLC30A10 | SLC30A10 | 3255 | 0.162 | 0.2335 | No |
| 19 | PARP8 | PARP8 | 3498 | 0.141 | 0.2327 | No |
| 20 | VIP | VIP | 3575 | 0.135 | 0.2384 | No |
| 21 | MSRB2 | MSRB2 | 3646 | 0.131 | 0.2440 | No |
| 22 | SGK3 | SGK3 | 3694 | 0.128 | 0.2504 | No |
| 23 | BCAS1 | BCAS1 | 3907 | 0.115 | 0.2492 | No |
| 24 | FUT9 | FUT9 | 4325 | 0.097 | 0.2383 | No |
| 25 | BASP1 | BASP1 | 4446 | 0.091 | 0.2393 | No |
| 26 | NSL1 | NSL1 | 4628 | 0.083 | 0.2372 | No |
| 27 | SH3GL3 | SH3GL3 | 5249 | 0.066 | 0.2160 | No |
| 28 | NANOS1 | NANOS1 | 5276 | 0.065 | 0.2192 | No |
| 29 | THTPA | THTPA | 5396 | 0.062 | 0.2183 | No |
| 30 | MGST3 | MGST3 | 6261 | 0.046 | 0.1857 | No |
| 31 | TTPA | TTPA | 6363 | 0.045 | 0.1845 | No |
| 32 | RND1 | RND1 | 6564 | 0.041 | 0.1789 | No |
| 33 | MMD | MMD | 6668 | 0.040 | 0.1773 | No |
| 34 | PRPSAP2 | PRPSAP2 | 7057 | 0.035 | 0.1636 | No |
| 35 | ATP5L | ATP5L | 7897 | 0.025 | 0.1306 | No |
| 36 | ANKRD46 | ANKRD46 | 8309 | 0.021 | 0.1151 | No |
| 37 | PDE6D | PDE6D | 8348 | 0.020 | 0.1148 | No |
| 38 | AGTPBP1 | AGTPBP1 | 8471 | 0.019 | 0.1110 | No |
| 39 | UROS | UROS | 8521 | 0.018 | 0.1102 | No |
| 40 | SEPP1 | SEPP1 | 8525 | 0.018 | 0.1113 | No |
| 41 | COX5B | COX5B | 8565 | 0.018 | 0.1108 | No |
| 42 | PEX19 | PEX19 | 8643 | 0.017 | 0.1088 | No |
| 43 | MDH1 | MDH1 | 8670 | 0.017 | 0.1089 | No |
| 44 | MAGEH1 | MAGEH1 | 8785 | 0.016 | 0.1052 | No |
| 45 | FAM49B | FAM49B | 8965 | 0.015 | 0.0988 | No |
| 46 | GABARAPL2 | GABARAPL2 | 9124 | 0.013 | 0.0931 | No |
| 47 | ADD3 | ADD3 | 9171 | 0.013 | 0.0921 | No |
| 48 | PGBD5 | PGBD5 | 9287 | 0.012 | 0.0881 | No |
| 49 | CUTC | CUTC | 9327 | 0.011 | 0.0872 | No |
| 50 | FBXO3 | FBXO3 | 9645 | 0.008 | 0.0747 | No |
| 51 | MAT2B | MAT2B | 9680 | 0.008 | 0.0739 | No |
| 52 | CAMK2G | CAMK2G | 9770 | 0.007 | 0.0707 | No |
| 53 | CALM2 | CALM2 | 9825 | 0.007 | 0.0689 | No |
| 54 | MRPL49 | MRPL49 | 9872 | 0.006 | 0.0674 | No |
| 55 | IMPA1 | IMPA1 | 9894 | 0.006 | 0.0670 | No |
| 56 | USP33 | USP33 | 10036 | 0.005 | 0.0615 | No |
| 57 | ANKS1B | ANKS1B | 10598 | 0.000 | 0.0384 | No |
| 58 | GUK1 | GUK1 | 10634 | 0.000 | 0.0370 | No |
| 59 | CDR1 | CDR1 | 12130 | 0.000 | -0.0246 | No |
| 60 | TSNAX | TSNAX | 15038 | -0.002 | -0.1442 | No |
| 61 | NDUFS3 | NDUFS3 | 15058 | -0.002 | -0.1449 | No |
| 62 | PEX11B | PEX11B | 15092 | -0.002 | -0.1461 | No |
| 63 | SPAST | SPAST | 15424 | -0.005 | -0.1594 | No |
| 64 | MORF4L2 | MORF4L2 | 15579 | -0.006 | -0.1653 | No |
| 65 | SEPW1 | SEPW1 | 15820 | -0.009 | -0.1746 | No |
| 66 | CRYZL1 | CRYZL1 | 15844 | -0.009 | -0.1750 | No |
| 67 | SH3GL2 | SH3GL2 | 16001 | -0.011 | -0.1807 | No |
| 68 | PPA1 | PPA1 | 16171 | -0.012 | -0.1869 | No |
| 69 | CRBN | CRBN | 16294 | -0.014 | -0.1910 | No |
| 70 | SIRT5 | SIRT5 | 16522 | -0.016 | -0.1994 | No |
| 71 | TTC1 | TTC1 | 16527 | -0.016 | -0.1985 | No |
| 72 | SAR1A | SAR1A | 16535 | -0.016 | -0.1977 | No |
| 73 | FHIT | FHIT | 16575 | -0.016 | -0.1983 | No |
| 74 | ENPP4 | ENPP4 | 16681 | -0.017 | -0.2015 | No |
| 75 | KCNK1 | KCNK1 | 16730 | -0.018 | -0.2023 | No |
| 76 | YPEL5 | YPEL5 | 16866 | -0.019 | -0.2066 | No |
| 77 | ROGDI | ROGDI | 16907 | -0.020 | -0.2069 | No |
| 78 | SNX11 | SNX11 | 17183 | -0.023 | -0.2167 | No |
| 79 | SCHIP1 | SCHIP1 | 17213 | -0.024 | -0.2164 | No |
| 80 | CDC42 | CDC42 | 17540 | -0.028 | -0.2280 | No |
| 81 | ATP5F1 | ATP5F1 | 17575 | -0.028 | -0.2276 | No |
| 82 | ACYP2 | ACYP2 | 17841 | -0.032 | -0.2364 | No |
| 83 | PPP2R5A | PPP2R5A | 18222 | -0.038 | -0.2496 | No |
| 84 | CASQ1 | CASQ1 | 18278 | -0.039 | -0.2493 | No |
| 85 | DHRS9 | DHRS9 | 18868 | -0.052 | -0.2702 | No |
| 86 | TPM3 | TPM3 | 18881 | -0.052 | -0.2673 | No |
| 87 | SLC16A7 | SLC16A7 | 19189 | -0.061 | -0.2760 | No |
| 88 | GNAI1 | GNAI1 | 19333 | -0.066 | -0.2776 | No |
| 89 | ATRNL1 | ATRNL1 | 19394 | -0.068 | -0.2756 | No |
| 90 | CCK | CCK | 19498 | -0.072 | -0.2752 | No |
| 91 | SERPINI1 | SERPINI1 | 19551 | -0.073 | -0.2725 | No |
| 92 | ACSL4 | ACSL4 | 19608 | -0.076 | -0.2699 | No |
| 93 | REPS2 | REPS2 | 19681 | -0.079 | -0.2677 | No |
| 94 | SLC31A2 | SLC31A2 | 19735 | -0.081 | -0.2647 | No |
| 95 | LYRM1 | LYRM1 | 19848 | -0.086 | -0.2636 | No |
| 96 | PLCL1 | PLCL1 | 20206 | -0.104 | -0.2716 | No |
| 97 | HPCAL4 | HPCAL4 | 20289 | -0.107 | -0.2680 | No |
| 98 | ARRB1 | ARRB1 | 20570 | -0.120 | -0.2717 | No |
| 99 | CPNE6 | CPNE6 | 20822 | -0.135 | -0.2733 | No |
| 100 | KCNJ3 | KCNJ3 | 21094 | -0.161 | -0.2740 | No |
| 101 | ANXA7 | ANXA7 | 21512 | -0.205 | -0.2778 | Yes |
| 102 | MYBPC1 | MYBPC1 | 21577 | -0.211 | -0.2667 | Yes |
| 103 | FEZF2 | FEZF2 | 21775 | -0.235 | -0.2595 | Yes |
| 104 | CHN1 | CHN1 | 22289 | -0.319 | -0.2599 | Yes |
| 105 | SNCG | SNCG | 23367 | -0.436 | -0.2759 | Yes |
| 106 | MBP | MBP | 23407 | -0.450 | -0.2482 | Yes |
| 107 | ANXA3 | ANXA3 | 23521 | -0.490 | -0.2209 | Yes |
| 108 | DYNC1I1 | DYNC1I1 | 23550 | -0.503 | -0.1893 | Yes |
| 109 | TMEM144 | TMEM144 | 23720 | -0.588 | -0.1580 | Yes |
| 110 | CRYM | CRYM | 23939 | -0.689 | -0.1221 | Yes |
| 111 | GPR22 | GPR22 | 23994 | -0.730 | -0.0768 | Yes |
| 112 | EVI2A | EVI2A | 24308 | -1.428 | 0.0033 | Yes |