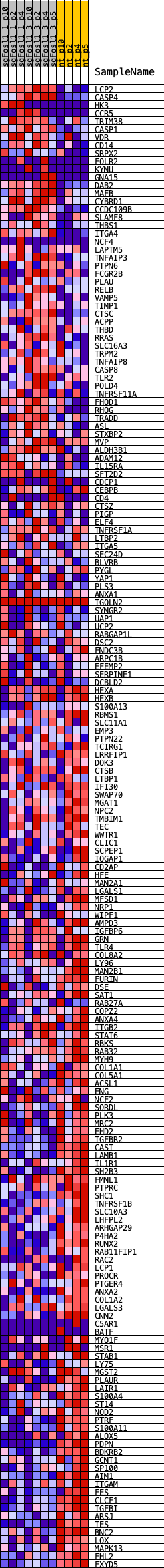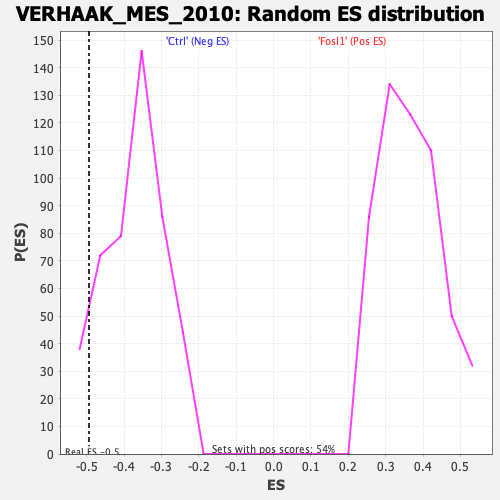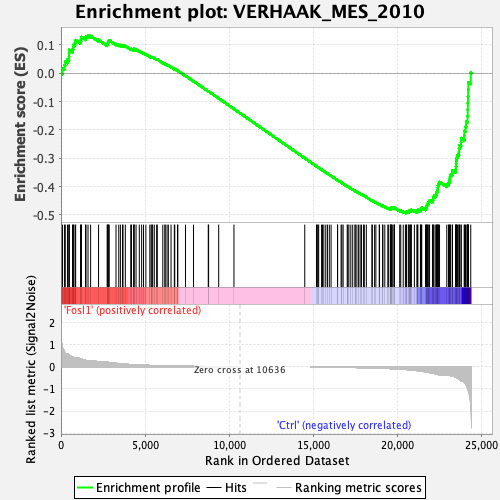
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | VERHAAK_MES_2010 |
| Enrichment Score (ES) | -0.4938699 |
| Normalized Enrichment Score (NES) | -1.3236738 |
| Nominal p-value | 0.07526882 |
| FDR q-value | 0.1591419 |
| FWER p-Value | 0.628 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LCP2 | LCP2 | 87 | 0.886 | 0.0171 | No |
| 2 | CASP4 | CASP4 | 204 | 0.693 | 0.0285 | No |
| 3 | HK3 | HK3 | 254 | 0.617 | 0.0409 | No |
| 4 | CCR5 | CCR5 | 384 | 0.582 | 0.0491 | No |
| 5 | TRIM38 | TRIM38 | 474 | 0.539 | 0.0581 | No |
| 6 | CASP1 | CASP1 | 480 | 0.536 | 0.0704 | No |
| 7 | VDR | VDR | 484 | 0.533 | 0.0827 | No |
| 8 | CD14 | CD14 | 684 | 0.440 | 0.0848 | No |
| 9 | SRPX2 | SRPX2 | 724 | 0.434 | 0.0933 | No |
| 10 | FOLR2 | FOLR2 | 752 | 0.430 | 0.1022 | No |
| 11 | KYNU | KYNU | 837 | 0.419 | 0.1085 | No |
| 12 | GNA15 | GNA15 | 872 | 0.416 | 0.1169 | No |
| 13 | DAB2 | DAB2 | 1160 | 0.365 | 0.1135 | No |
| 14 | MAFB | MAFB | 1188 | 0.358 | 0.1208 | No |
| 15 | CYBRD1 | CYBRD1 | 1216 | 0.351 | 0.1279 | No |
| 16 | CCDC109B | CCDC109B | 1459 | 0.295 | 0.1248 | No |
| 17 | SLAMF8 | SLAMF8 | 1485 | 0.290 | 0.1305 | No |
| 18 | THBS1 | THBS1 | 1605 | 0.272 | 0.1319 | No |
| 19 | ITGA4 | ITGA4 | 1760 | 0.254 | 0.1315 | No |
| 20 | NCF4 | NCF4 | 2232 | 0.245 | 0.1178 | No |
| 21 | LAPTM5 | LAPTM5 | 2742 | 0.215 | 0.1018 | No |
| 22 | TNFAIP3 | TNFAIP3 | 2772 | 0.211 | 0.1055 | No |
| 23 | PTPN6 | PTPN6 | 2801 | 0.207 | 0.1092 | No |
| 24 | FCGR2B | FCGR2B | 2808 | 0.206 | 0.1137 | No |
| 25 | PLAU | PLAU | 2866 | 0.199 | 0.1160 | No |
| 26 | RELB | RELB | 3268 | 0.161 | 0.1032 | No |
| 27 | VAMP5 | VAMP5 | 3432 | 0.147 | 0.0999 | No |
| 28 | TIMP1 | TIMP1 | 3528 | 0.139 | 0.0992 | No |
| 29 | CTSC | CTSC | 3655 | 0.131 | 0.0971 | No |
| 30 | ACPP | ACPP | 3683 | 0.129 | 0.0990 | No |
| 31 | THBD | THBD | 3821 | 0.119 | 0.0961 | No |
| 32 | RRAS | RRAS | 4145 | 0.105 | 0.0852 | No |
| 33 | SLC16A3 | SLC16A3 | 4187 | 0.103 | 0.0859 | No |
| 34 | TRPM2 | TRPM2 | 4323 | 0.097 | 0.0826 | No |
| 35 | TNFAIP8 | TNFAIP8 | 4331 | 0.096 | 0.0846 | No |
| 36 | CASP8 | CASP8 | 4344 | 0.096 | 0.0863 | No |
| 37 | TLR2 | TLR2 | 4459 | 0.090 | 0.0837 | No |
| 38 | POLD4 | POLD4 | 4652 | 0.083 | 0.0777 | No |
| 39 | TNFRSF11A | TNFRSF11A | 4792 | 0.078 | 0.0738 | No |
| 40 | FHOD1 | FHOD1 | 4903 | 0.075 | 0.0710 | No |
| 41 | RHOG | RHOG | 5042 | 0.071 | 0.0670 | No |
| 42 | TRADD | TRADD | 5278 | 0.065 | 0.0588 | No |
| 43 | ASL | ASL | 5355 | 0.063 | 0.0571 | No |
| 44 | STXBP2 | STXBP2 | 5402 | 0.062 | 0.0566 | No |
| 45 | MVP | MVP | 5446 | 0.061 | 0.0563 | No |
| 46 | ALDH3B1 | ALDH3B1 | 5542 | 0.059 | 0.0538 | No |
| 47 | ADAM12 | ADAM12 | 5685 | 0.056 | 0.0492 | No |
| 48 | IL15RA | IL15RA | 5720 | 0.056 | 0.0491 | No |
| 49 | SFT2D2 | SFT2D2 | 6048 | 0.050 | 0.0368 | No |
| 50 | CDCP1 | CDCP1 | 6176 | 0.048 | 0.0326 | No |
| 51 | CEBPB | CEBPB | 6185 | 0.048 | 0.0334 | No |
| 52 | CD4 | CD4 | 6299 | 0.046 | 0.0298 | No |
| 53 | CTSZ | CTSZ | 6378 | 0.044 | 0.0276 | No |
| 54 | PIGP | PIGP | 6538 | 0.042 | 0.0220 | No |
| 55 | ELF4 | ELF4 | 6733 | 0.039 | 0.0149 | No |
| 56 | TNFRSF1A | TNFRSF1A | 6759 | 0.039 | 0.0148 | No |
| 57 | LTBP2 | LTBP2 | 6925 | 0.036 | 0.0088 | No |
| 58 | ITGA5 | ITGA5 | 6940 | 0.036 | 0.0091 | No |
| 59 | SEC24D | SEC24D | 7396 | 0.031 | -0.0090 | No |
| 60 | BLVRB | BLVRB | 7872 | 0.025 | -0.0280 | No |
| 61 | PYGL | PYGL | 8749 | 0.017 | -0.0638 | No |
| 62 | YAP1 | YAP1 | 8771 | 0.016 | -0.0643 | No |
| 63 | PLS3 | PLS3 | 9369 | 0.011 | -0.0887 | No |
| 64 | ANXA1 | ANXA1 | 10276 | 0.003 | -0.1261 | No |
| 65 | TGOLN2 | TGOLN2 | 14483 | 0.000 | -0.2998 | No |
| 66 | SYNGR2 | SYNGR2 | 15178 | -0.003 | -0.3284 | No |
| 67 | UAP1 | UAP1 | 15225 | -0.004 | -0.3303 | No |
| 68 | UCP2 | UCP2 | 15270 | -0.004 | -0.3320 | No |
| 69 | RABGAP1L | RABGAP1L | 15310 | -0.004 | -0.3335 | No |
| 70 | DSC2 | DSC2 | 15482 | -0.006 | -0.3404 | No |
| 71 | FNDC3B | FNDC3B | 15538 | -0.006 | -0.3426 | No |
| 72 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.3456 | No |
| 73 | EFEMP2 | EFEMP2 | 15731 | -0.008 | -0.3501 | No |
| 74 | SERPINE1 | SERPINE1 | 15830 | -0.009 | -0.3539 | No |
| 75 | DCBLD2 | DCBLD2 | 15941 | -0.010 | -0.3583 | No |
| 76 | HEXA | HEXA | 16046 | -0.011 | -0.3623 | No |
| 77 | HEXB | HEXB | 16429 | -0.015 | -0.3777 | No |
| 78 | S100A13 | S100A13 | 16435 | -0.015 | -0.3776 | No |
| 79 | RBMS1 | RBMS1 | 16641 | -0.017 | -0.3857 | No |
| 80 | SLC11A1 | SLC11A1 | 16688 | -0.017 | -0.3872 | No |
| 81 | EMP3 | EMP3 | 16762 | -0.018 | -0.3897 | No |
| 82 | PTPN22 | PTPN22 | 17005 | -0.021 | -0.3992 | No |
| 83 | TCIRG1 | TCIRG1 | 17040 | -0.021 | -0.4002 | No |
| 84 | LRRFIP1 | LRRFIP1 | 17105 | -0.022 | -0.4023 | No |
| 85 | DOK3 | DOK3 | 17219 | -0.024 | -0.4064 | No |
| 86 | CTSB | CTSB | 17332 | -0.025 | -0.4104 | No |
| 87 | LTBP1 | LTBP1 | 17471 | -0.027 | -0.4155 | No |
| 88 | IFI30 | IFI30 | 17476 | -0.027 | -0.4150 | No |
| 89 | SWAP70 | SWAP70 | 17580 | -0.028 | -0.4186 | No |
| 90 | MGAT1 | MGAT1 | 17682 | -0.030 | -0.4221 | No |
| 91 | NPC2 | NPC2 | 17773 | -0.031 | -0.4251 | No |
| 92 | TMBIM1 | TMBIM1 | 17856 | -0.032 | -0.4278 | No |
| 93 | TEC | TEC | 17977 | -0.034 | -0.4319 | No |
| 94 | WWTR1 | WWTR1 | 18020 | -0.035 | -0.4328 | No |
| 95 | CLIC1 | CLIC1 | 18140 | -0.037 | -0.4369 | No |
| 96 | SCPEP1 | SCPEP1 | 18472 | -0.043 | -0.4496 | No |
| 97 | IQGAP1 | IQGAP1 | 18498 | -0.043 | -0.4496 | No |
| 98 | CD2AP | CD2AP | 18633 | -0.046 | -0.4540 | No |
| 99 | HFE | HFE | 18708 | -0.048 | -0.4560 | No |
| 100 | MAN2A1 | MAN2A1 | 18907 | -0.053 | -0.4629 | No |
| 101 | LGALS1 | LGALS1 | 18926 | -0.053 | -0.4624 | No |
| 102 | MFSD1 | MFSD1 | 19110 | -0.058 | -0.4686 | No |
| 103 | NRP1 | NRP1 | 19122 | -0.059 | -0.4677 | No |
| 104 | WIPF1 | WIPF1 | 19239 | -0.063 | -0.4710 | No |
| 105 | AMPD3 | AMPD3 | 19418 | -0.069 | -0.4768 | No |
| 106 | IGFBP6 | IGFBP6 | 19469 | -0.071 | -0.4772 | No |
| 107 | GRN | GRN | 19573 | -0.074 | -0.4797 | No |
| 108 | TLR4 | TLR4 | 19589 | -0.075 | -0.4786 | No |
| 109 | COL8A2 | COL8A2 | 19604 | -0.076 | -0.4774 | No |
| 110 | LY96 | LY96 | 19617 | -0.076 | -0.4761 | No |
| 111 | MAN2B1 | MAN2B1 | 19621 | -0.076 | -0.4744 | No |
| 112 | FURIN | FURIN | 19684 | -0.079 | -0.4752 | No |
| 113 | DSE | DSE | 19781 | -0.083 | -0.4772 | No |
| 114 | SAT1 | SAT1 | 19808 | -0.084 | -0.4763 | No |
| 115 | RAB27A | RAB27A | 19809 | -0.084 | -0.4743 | No |
| 116 | COPZ2 | COPZ2 | 20125 | -0.100 | -0.4850 | No |
| 117 | ANXA4 | ANXA4 | 20186 | -0.103 | -0.4851 | No |
| 118 | ITGB2 | ITGB2 | 20333 | -0.108 | -0.4886 | No |
| 119 | STAT6 | STAT6 | 20462 | -0.115 | -0.4912 | Yes |
| 120 | RBKS | RBKS | 20507 | -0.117 | -0.4903 | Yes |
| 121 | RAB32 | RAB32 | 20529 | -0.119 | -0.4883 | Yes |
| 122 | MYH9 | MYH9 | 20636 | -0.124 | -0.4898 | Yes |
| 123 | COL1A1 | COL1A1 | 20681 | -0.127 | -0.4887 | Yes |
| 124 | COL5A1 | COL5A1 | 20683 | -0.127 | -0.4857 | Yes |
| 125 | ACSL1 | ACSL1 | 20761 | -0.131 | -0.4858 | Yes |
| 126 | ENG | ENG | 20788 | -0.133 | -0.4838 | Yes |
| 127 | NCF2 | NCF2 | 20814 | -0.134 | -0.4817 | Yes |
| 128 | SQRDL | SQRDL | 20995 | -0.151 | -0.4856 | Yes |
| 129 | PLK3 | PLK3 | 21148 | -0.165 | -0.4880 | Yes |
| 130 | MRC2 | MRC2 | 21173 | -0.168 | -0.4851 | Yes |
| 131 | EHD2 | EHD2 | 21204 | -0.171 | -0.4824 | Yes |
| 132 | TGFBR2 | TGFBR2 | 21354 | -0.188 | -0.4841 | Yes |
| 133 | CAST | CAST | 21363 | -0.190 | -0.4800 | Yes |
| 134 | LAMB1 | LAMB1 | 21416 | -0.195 | -0.4776 | Yes |
| 135 | IL1R1 | IL1R1 | 21433 | -0.197 | -0.4737 | Yes |
| 136 | SH2B3 | SH2B3 | 21658 | -0.219 | -0.4778 | Yes |
| 137 | FMNL1 | FMNL1 | 21696 | -0.223 | -0.4741 | Yes |
| 138 | PTPRC | PTPRC | 21711 | -0.226 | -0.4694 | Yes |
| 139 | SHC1 | SHC1 | 21741 | -0.230 | -0.4653 | Yes |
| 140 | TNFRSF1B | TNFRSF1B | 21769 | -0.234 | -0.4609 | Yes |
| 141 | SLC10A3 | SLC10A3 | 21813 | -0.241 | -0.4571 | Yes |
| 142 | LHFPL2 | LHFPL2 | 21860 | -0.248 | -0.4532 | Yes |
| 143 | ARHGAP29 | ARHGAP29 | 21898 | -0.254 | -0.4488 | Yes |
| 144 | P4HA2 | P4HA2 | 22070 | -0.281 | -0.4493 | Yes |
| 145 | RUNX2 | RUNX2 | 22102 | -0.286 | -0.4439 | Yes |
| 146 | RAB11FIP1 | RAB11FIP1 | 22112 | -0.287 | -0.4375 | Yes |
| 147 | RAC2 | RAC2 | 22161 | -0.295 | -0.4326 | Yes |
| 148 | LCP1 | LCP1 | 22265 | -0.313 | -0.4296 | Yes |
| 149 | PROCR | PROCR | 22311 | -0.323 | -0.4239 | Yes |
| 150 | PTGER4 | PTGER4 | 22332 | -0.329 | -0.4170 | Yes |
| 151 | ANXA2 | ANXA2 | 22374 | -0.334 | -0.4109 | Yes |
| 152 | COL1A2 | COL1A2 | 22387 | -0.337 | -0.4035 | Yes |
| 153 | LGALS3 | LGALS3 | 22395 | -0.340 | -0.3959 | Yes |
| 154 | CNN2 | CNN2 | 22435 | -0.347 | -0.3894 | Yes |
| 155 | C5AR1 | C5AR1 | 22491 | -0.352 | -0.3834 | Yes |
| 156 | BATF | BATF | 22921 | -0.358 | -0.3928 | Yes |
| 157 | MYO1F | MYO1F | 23021 | -0.366 | -0.3883 | Yes |
| 158 | MSR1 | MSR1 | 23060 | -0.373 | -0.3812 | Yes |
| 159 | STAB1 | STAB1 | 23094 | -0.379 | -0.3737 | Yes |
| 160 | LY75 | LY75 | 23111 | -0.385 | -0.3654 | Yes |
| 161 | MGST2 | MGST2 | 23149 | -0.394 | -0.3577 | Yes |
| 162 | PLAUR | PLAUR | 23251 | -0.408 | -0.3524 | Yes |
| 163 | LAIR1 | LAIR1 | 23256 | -0.409 | -0.3430 | Yes |
| 164 | S100A4 | S100A4 | 23442 | -0.464 | -0.3398 | Yes |
| 165 | ST14 | ST14 | 23465 | -0.472 | -0.3296 | Yes |
| 166 | NOD2 | NOD2 | 23475 | -0.474 | -0.3189 | Yes |
| 167 | PTRF | PTRF | 23482 | -0.476 | -0.3081 | Yes |
| 168 | S100A11 | S100A11 | 23510 | -0.487 | -0.2978 | Yes |
| 169 | ALOX5 | ALOX5 | 23563 | -0.510 | -0.2880 | Yes |
| 170 | PDPN | PDPN | 23644 | -0.542 | -0.2787 | Yes |
| 171 | BDKRB2 | BDKRB2 | 23649 | -0.544 | -0.2661 | Yes |
| 172 | GCNT1 | GCNT1 | 23676 | -0.557 | -0.2542 | Yes |
| 173 | SP100 | SP100 | 23768 | -0.624 | -0.2434 | Yes |
| 174 | AIM1 | AIM1 | 23776 | -0.633 | -0.2289 | Yes |
| 175 | ITGAM | ITGAM | 23961 | -0.701 | -0.2201 | Yes |
| 176 | FES | FES | 23965 | -0.703 | -0.2038 | Yes |
| 177 | CLCF1 | CLCF1 | 24034 | -0.767 | -0.1887 | Yes |
| 178 | TGFBI | TGFBI | 24073 | -0.818 | -0.1711 | Yes |
| 179 | ARSJ | ARSJ | 24152 | -0.971 | -0.1517 | Yes |
| 180 | TES | TES | 24165 | -1.003 | -0.1287 | Yes |
| 181 | BNC2 | BNC2 | 24170 | -1.012 | -0.1053 | Yes |
| 182 | LOX | LOX | 24181 | -1.034 | -0.0815 | Yes |
| 183 | MAPK13 | MAPK13 | 24194 | -1.059 | -0.0573 | Yes |
| 184 | FHL2 | FHL2 | 24205 | -1.090 | -0.0322 | Yes |
| 185 | FXYD5 | FXYD5 | 24345 | -1.702 | 0.0018 | Yes |