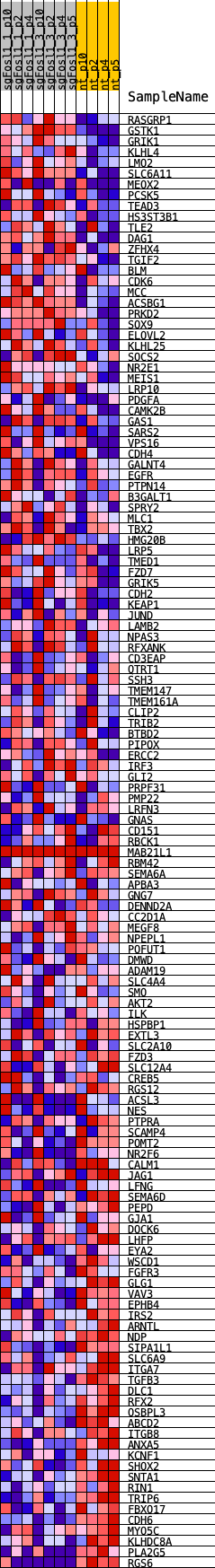
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | VERHAAK_CL_2010 |
| Enrichment Score (ES) | -0.3692746 |
| Normalized Enrichment Score (NES) | -1.4670454 |
| Nominal p-value | 0.014675053 |
| FDR q-value | 0.07848638 |
| FWER p-Value | 0.282 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RASGRP1 | RASGRP1 | 91 | 0.862 | 0.0512 | No |
| 2 | GSTK1 | GSTK1 | 481 | 0.535 | 0.0693 | No |
| 3 | GRIK1 | GRIK1 | 494 | 0.526 | 0.1024 | No |
| 4 | KLHL4 | KLHL4 | 639 | 0.457 | 0.1256 | No |
| 5 | LMO2 | LMO2 | 1484 | 0.290 | 0.1093 | No |
| 6 | SLC6A11 | SLC6A11 | 1647 | 0.264 | 0.1195 | No |
| 7 | MEOX2 | MEOX2 | 2727 | 0.218 | 0.0889 | No |
| 8 | PCSK5 | PCSK5 | 2753 | 0.214 | 0.1015 | No |
| 9 | TEAD3 | TEAD3 | 3573 | 0.135 | 0.0763 | No |
| 10 | HS3ST3B1 | HS3ST3B1 | 3652 | 0.131 | 0.0815 | No |
| 11 | TLE2 | TLE2 | 3879 | 0.117 | 0.0796 | No |
| 12 | DAG1 | DAG1 | 3934 | 0.114 | 0.0847 | No |
| 13 | ZFHX4 | ZFHX4 | 3967 | 0.112 | 0.0905 | No |
| 14 | TGIF2 | TGIF2 | 4359 | 0.095 | 0.0805 | No |
| 15 | BLM | BLM | 4591 | 0.085 | 0.0764 | No |
| 16 | CDK6 | CDK6 | 4730 | 0.080 | 0.0758 | No |
| 17 | MCC | MCC | 4844 | 0.077 | 0.0760 | No |
| 18 | ACSBG1 | ACSBG1 | 5032 | 0.072 | 0.0729 | No |
| 19 | PRKD2 | PRKD2 | 5080 | 0.070 | 0.0754 | No |
| 20 | SOX9 | SOX9 | 5082 | 0.070 | 0.0799 | No |
| 21 | ELOVL2 | ELOVL2 | 5216 | 0.067 | 0.0786 | No |
| 22 | KLHL25 | KLHL25 | 5271 | 0.065 | 0.0806 | No |
| 23 | SOCS2 | SOCS2 | 5300 | 0.064 | 0.0835 | No |
| 24 | NR2E1 | NR2E1 | 5406 | 0.062 | 0.0831 | No |
| 25 | MEIS1 | MEIS1 | 6012 | 0.050 | 0.0614 | No |
| 26 | LRP10 | LRP10 | 6021 | 0.050 | 0.0643 | No |
| 27 | PDGFA | PDGFA | 6104 | 0.049 | 0.0640 | No |
| 28 | CAMK2B | CAMK2B | 6257 | 0.046 | 0.0607 | No |
| 29 | GAS1 | GAS1 | 6287 | 0.046 | 0.0624 | No |
| 30 | SARS2 | SARS2 | 6432 | 0.043 | 0.0593 | No |
| 31 | VPS16 | VPS16 | 6540 | 0.042 | 0.0575 | No |
| 32 | CDH4 | CDH4 | 6583 | 0.041 | 0.0584 | No |
| 33 | GALNT4 | GALNT4 | 6630 | 0.040 | 0.0591 | No |
| 34 | EGFR | EGFR | 6651 | 0.040 | 0.0609 | No |
| 35 | PTPN14 | PTPN14 | 6727 | 0.039 | 0.0603 | No |
| 36 | B3GALT1 | B3GALT1 | 6932 | 0.036 | 0.0542 | No |
| 37 | SPRY2 | SPRY2 | 6975 | 0.036 | 0.0547 | No |
| 38 | MLC1 | MLC1 | 7007 | 0.035 | 0.0557 | No |
| 39 | TBX2 | TBX2 | 7148 | 0.034 | 0.0521 | No |
| 40 | HMG20B | HMG20B | 7249 | 0.033 | 0.0501 | No |
| 41 | LRP5 | LRP5 | 7328 | 0.032 | 0.0489 | No |
| 42 | TMED1 | TMED1 | 7409 | 0.031 | 0.0475 | No |
| 43 | FZD7 | FZD7 | 7513 | 0.029 | 0.0451 | No |
| 44 | GRIK5 | GRIK5 | 7519 | 0.029 | 0.0468 | No |
| 45 | CDH2 | CDH2 | 7602 | 0.028 | 0.0452 | No |
| 46 | KEAP1 | KEAP1 | 7784 | 0.026 | 0.0394 | No |
| 47 | JUND | JUND | 7816 | 0.026 | 0.0398 | No |
| 48 | LAMB2 | LAMB2 | 7819 | 0.026 | 0.0414 | No |
| 49 | NPAS3 | NPAS3 | 7884 | 0.025 | 0.0404 | No |
| 50 | RFXANK | RFXANK | 7932 | 0.025 | 0.0400 | No |
| 51 | CD3EAP | CD3EAP | 8646 | 0.017 | 0.0117 | No |
| 52 | QTRT1 | QTRT1 | 8759 | 0.016 | 0.0081 | No |
| 53 | SSH3 | SSH3 | 8848 | 0.016 | 0.0055 | No |
| 54 | TMEM147 | TMEM147 | 8881 | 0.015 | 0.0052 | No |
| 55 | TMEM161A | TMEM161A | 8968 | 0.015 | 0.0026 | No |
| 56 | CLIP2 | CLIP2 | 9027 | 0.014 | 0.0011 | No |
| 57 | TRIB2 | TRIB2 | 9055 | 0.014 | 0.0008 | No |
| 58 | BTBD2 | BTBD2 | 9056 | 0.014 | 0.0017 | No |
| 59 | PIPOX | PIPOX | 9202 | 0.012 | -0.0035 | No |
| 60 | ERCC2 | ERCC2 | 9406 | 0.011 | -0.0112 | No |
| 61 | IRF3 | IRF3 | 9844 | 0.007 | -0.0288 | No |
| 62 | GLI2 | GLI2 | 9871 | 0.006 | -0.0294 | No |
| 63 | PRPF31 | PRPF31 | 10317 | 0.003 | -0.0476 | No |
| 64 | PMP22 | PMP22 | 10332 | 0.003 | -0.0480 | No |
| 65 | LRFN3 | LRFN3 | 10407 | 0.002 | -0.0509 | No |
| 66 | GNAS | GNAS | 10525 | 0.001 | -0.0557 | No |
| 67 | CD151 | CD151 | 10556 | 0.001 | -0.0569 | No |
| 68 | RBCK1 | RBCK1 | 10614 | 0.000 | -0.0592 | No |
| 69 | MAB21L1 | MAB21L1 | 13233 | 0.000 | -0.1672 | No |
| 70 | RBM42 | RBM42 | 14856 | -0.000 | -0.2340 | No |
| 71 | SEMA6A | SEMA6A | 14984 | -0.001 | -0.2392 | No |
| 72 | APBA3 | APBA3 | 14992 | -0.001 | -0.2394 | No |
| 73 | GNG7 | GNG7 | 15012 | -0.002 | -0.2401 | No |
| 74 | DENND2A | DENND2A | 15033 | -0.002 | -0.2408 | No |
| 75 | CC2D1A | CC2D1A | 15157 | -0.003 | -0.2457 | No |
| 76 | MEGF8 | MEGF8 | 15213 | -0.003 | -0.2477 | No |
| 77 | NPEPL1 | NPEPL1 | 15487 | -0.006 | -0.2586 | No |
| 78 | POFUT1 | POFUT1 | 15550 | -0.006 | -0.2608 | No |
| 79 | DMWD | DMWD | 15776 | -0.008 | -0.2695 | No |
| 80 | ADAM19 | ADAM19 | 15961 | -0.010 | -0.2764 | No |
| 81 | SLC4A4 | SLC4A4 | 16396 | -0.015 | -0.2934 | No |
| 82 | SMO | SMO | 16404 | -0.015 | -0.2928 | No |
| 83 | AKT2 | AKT2 | 16496 | -0.016 | -0.2955 | No |
| 84 | ILK | ILK | 16632 | -0.017 | -0.3000 | No |
| 85 | HSPBP1 | HSPBP1 | 16652 | -0.017 | -0.2997 | No |
| 86 | EXTL3 | EXTL3 | 16675 | -0.017 | -0.2995 | No |
| 87 | SLC2A10 | SLC2A10 | 16896 | -0.020 | -0.3073 | No |
| 88 | FZD3 | FZD3 | 16912 | -0.020 | -0.3067 | No |
| 89 | SLC12A4 | SLC12A4 | 17032 | -0.021 | -0.3102 | No |
| 90 | CREB5 | CREB5 | 17544 | -0.028 | -0.3295 | No |
| 91 | RGS12 | RGS12 | 17685 | -0.030 | -0.3334 | No |
| 92 | ACSL3 | ACSL3 | 17708 | -0.030 | -0.3324 | No |
| 93 | NES | NES | 17881 | -0.033 | -0.3374 | No |
| 94 | PTPRA | PTPRA | 17926 | -0.033 | -0.3371 | No |
| 95 | SCAMP4 | SCAMP4 | 18091 | -0.036 | -0.3415 | No |
| 96 | POMT2 | POMT2 | 18099 | -0.036 | -0.3395 | No |
| 97 | NR2F6 | NR2F6 | 18216 | -0.038 | -0.3419 | No |
| 98 | CALM1 | CALM1 | 18375 | -0.041 | -0.3457 | No |
| 99 | JAG1 | JAG1 | 18825 | -0.051 | -0.3610 | No |
| 100 | LFNG | LFNG | 19026 | -0.056 | -0.3657 | Yes |
| 101 | SEMA6D | SEMA6D | 19040 | -0.056 | -0.3627 | Yes |
| 102 | PEPD | PEPD | 19059 | -0.057 | -0.3598 | Yes |
| 103 | GJA1 | GJA1 | 19177 | -0.061 | -0.3607 | Yes |
| 104 | DOCK6 | DOCK6 | 19366 | -0.067 | -0.3642 | Yes |
| 105 | LHFP | LHFP | 19437 | -0.070 | -0.3626 | Yes |
| 106 | EYA2 | EYA2 | 19479 | -0.071 | -0.3598 | Yes |
| 107 | WSCD1 | WSCD1 | 19539 | -0.073 | -0.3576 | Yes |
| 108 | FGFR3 | FGFR3 | 19566 | -0.074 | -0.3539 | Yes |
| 109 | GLG1 | GLG1 | 19687 | -0.079 | -0.3538 | Yes |
| 110 | VAV3 | VAV3 | 19782 | -0.083 | -0.3524 | Yes |
| 111 | EPHB4 | EPHB4 | 19784 | -0.083 | -0.3471 | Yes |
| 112 | IRS2 | IRS2 | 19841 | -0.086 | -0.3439 | Yes |
| 113 | ARNTL | ARNTL | 19932 | -0.090 | -0.3419 | Yes |
| 114 | NDP | NDP | 19970 | -0.092 | -0.3375 | Yes |
| 115 | SIPA1L1 | SIPA1L1 | 20025 | -0.094 | -0.3337 | Yes |
| 116 | SLC6A9 | SLC6A9 | 20384 | -0.111 | -0.3414 | Yes |
| 117 | ITGA7 | ITGA7 | 20879 | -0.139 | -0.3529 | Yes |
| 118 | TGFB3 | TGFB3 | 20922 | -0.143 | -0.3455 | Yes |
| 119 | DLC1 | DLC1 | 20939 | -0.145 | -0.3370 | Yes |
| 120 | RFX2 | RFX2 | 21002 | -0.152 | -0.3298 | Yes |
| 121 | OSBPL3 | OSBPL3 | 21074 | -0.159 | -0.3226 | Yes |
| 122 | ABCD2 | ABCD2 | 21118 | -0.163 | -0.3140 | Yes |
| 123 | ITGB8 | ITGB8 | 21155 | -0.166 | -0.3049 | Yes |
| 124 | ANXA5 | ANXA5 | 21429 | -0.196 | -0.3036 | Yes |
| 125 | KCNF1 | KCNF1 | 21655 | -0.219 | -0.2989 | Yes |
| 126 | SHOX2 | SHOX2 | 21794 | -0.237 | -0.2895 | Yes |
| 127 | SNTA1 | SNTA1 | 22055 | -0.278 | -0.2825 | Yes |
| 128 | RIN1 | RIN1 | 22120 | -0.288 | -0.2667 | Yes |
| 129 | TRIP6 | TRIP6 | 22201 | -0.303 | -0.2507 | Yes |
| 130 | FBXO17 | FBXO17 | 23057 | -0.373 | -0.2621 | Yes |
| 131 | CDH6 | CDH6 | 23760 | -0.620 | -0.2515 | Yes |
| 132 | MYO5C | MYO5C | 23863 | -0.642 | -0.2147 | Yes |
| 133 | KLHDC8A | KLHDC8A | 24031 | -0.764 | -0.1728 | Yes |
| 134 | PLA2G5 | PLA2G5 | 24243 | -1.172 | -0.1067 | Yes |
| 135 | RGS6 | RGS6 | 24354 | -1.765 | 0.0014 | Yes |