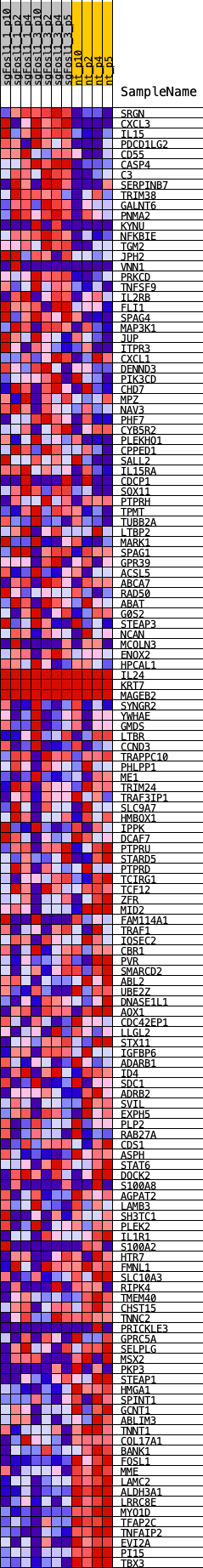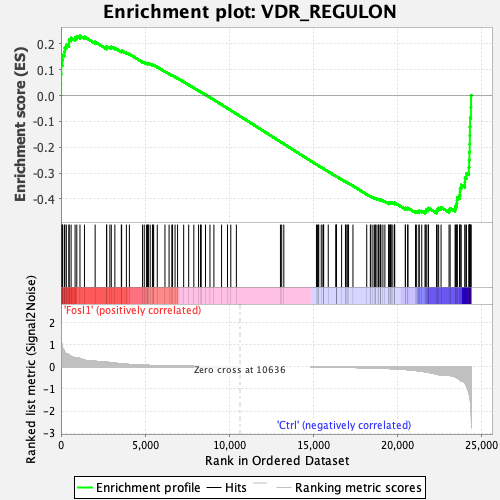
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | VDR_REGULON |
| Enrichment Score (ES) | -0.4548373 |
| Normalized Enrichment Score (NES) | -1.2117401 |
| Nominal p-value | 0.14851485 |
| FDR q-value | 0.299065 |
| FWER p-Value | 0.892 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SRGN | SRGN | 4 | 1.750 | 0.0430 | No |
| 2 | CXCL3 | CXCL3 | 6 | 1.622 | 0.0829 | No |
| 3 | IL15 | IL15 | 15 | 1.425 | 0.1177 | No |
| 4 | PDCD1LG2 | PDCD1LG2 | 78 | 0.906 | 0.1375 | No |
| 5 | CD55 | CD55 | 100 | 0.841 | 0.1573 | No |
| 6 | CASP4 | CASP4 | 204 | 0.693 | 0.1702 | No |
| 7 | C3 | C3 | 228 | 0.660 | 0.1855 | No |
| 8 | SERPINB7 | SERPINB7 | 317 | 0.593 | 0.1965 | No |
| 9 | TRIM38 | TRIM38 | 474 | 0.539 | 0.2033 | No |
| 10 | GALNT6 | GALNT6 | 478 | 0.537 | 0.2164 | No |
| 11 | PNMA2 | PNMA2 | 600 | 0.472 | 0.2231 | No |
| 12 | KYNU | KYNU | 837 | 0.419 | 0.2237 | No |
| 13 | NFKBIE | NFKBIE | 939 | 0.413 | 0.2297 | No |
| 14 | TGM2 | TGM2 | 1127 | 0.375 | 0.2313 | No |
| 15 | JPH2 | JPH2 | 1394 | 0.307 | 0.2279 | No |
| 16 | VNN1 | VNN1 | 2029 | 0.250 | 0.2079 | No |
| 17 | PRKCD | PRKCD | 2711 | 0.220 | 0.1852 | No |
| 18 | TNFSF9 | TNFSF9 | 2712 | 0.220 | 0.1906 | No |
| 19 | IL2RB | IL2RB | 2904 | 0.195 | 0.1876 | No |
| 20 | FLI1 | FLI1 | 2988 | 0.187 | 0.1887 | No |
| 21 | SPAG4 | SPAG4 | 3200 | 0.167 | 0.1842 | No |
| 22 | MAP3K1 | MAP3K1 | 3584 | 0.134 | 0.1717 | No |
| 23 | JUP | JUP | 3603 | 0.133 | 0.1742 | No |
| 24 | ITPR3 | ITPR3 | 3885 | 0.116 | 0.1655 | No |
| 25 | CXCL1 | CXCL1 | 4063 | 0.108 | 0.1609 | No |
| 26 | DENND3 | DENND3 | 4845 | 0.077 | 0.1305 | No |
| 27 | PIK3CD | PIK3CD | 4946 | 0.074 | 0.1282 | No |
| 28 | CHD7 | CHD7 | 5075 | 0.070 | 0.1247 | No |
| 29 | MPZ | MPZ | 5142 | 0.069 | 0.1237 | No |
| 30 | NAV3 | NAV3 | 5166 | 0.068 | 0.1244 | No |
| 31 | PHF7 | PHF7 | 5191 | 0.067 | 0.1250 | No |
| 32 | CYB5R2 | CYB5R2 | 5299 | 0.064 | 0.1222 | No |
| 33 | PLEKHO1 | PLEKHO1 | 5433 | 0.062 | 0.1183 | No |
| 34 | CPPED1 | CPPED1 | 5471 | 0.061 | 0.1182 | No |
| 35 | SALL2 | SALL2 | 5505 | 0.060 | 0.1184 | No |
| 36 | IL15RA | IL15RA | 5720 | 0.056 | 0.1109 | No |
| 37 | CDCP1 | CDCP1 | 6176 | 0.048 | 0.0933 | No |
| 38 | SOX11 | SOX11 | 6421 | 0.044 | 0.0843 | No |
| 39 | PTPRH | PTPRH | 6586 | 0.041 | 0.0786 | No |
| 40 | TPMT | TPMT | 6628 | 0.040 | 0.0779 | No |
| 41 | TUBB2A | TUBB2A | 6776 | 0.038 | 0.0728 | No |
| 42 | LTBP2 | LTBP2 | 6925 | 0.036 | 0.0676 | No |
| 43 | MARK1 | MARK1 | 7288 | 0.032 | 0.0534 | No |
| 44 | SPAG1 | SPAG1 | 7587 | 0.028 | 0.0418 | No |
| 45 | GPR39 | GPR39 | 7892 | 0.025 | 0.0299 | No |
| 46 | ACSL5 | ACSL5 | 8173 | 0.022 | 0.0189 | No |
| 47 | ABCA7 | ABCA7 | 8289 | 0.021 | 0.0147 | No |
| 48 | RAD50 | RAD50 | 8330 | 0.020 | 0.0135 | No |
| 49 | ABAT | ABAT | 8586 | 0.018 | 0.0034 | No |
| 50 | G0S2 | G0S2 | 8851 | 0.016 | -0.0071 | No |
| 51 | STEAP3 | STEAP3 | 9084 | 0.013 | -0.0163 | No |
| 52 | NCAN | NCAN | 9540 | 0.010 | -0.0348 | No |
| 53 | MCOLN3 | MCOLN3 | 9892 | 0.006 | -0.0491 | No |
| 54 | ENOX2 | ENOX2 | 10088 | 0.005 | -0.0571 | No |
| 55 | HPCAL1 | HPCAL1 | 10419 | 0.002 | -0.0706 | No |
| 56 | IL24 | IL24 | 13038 | 0.000 | -0.1786 | No |
| 57 | KRT7 | KRT7 | 13112 | 0.000 | -0.1816 | No |
| 58 | MAGEB2 | MAGEB2 | 13239 | 0.000 | -0.1868 | No |
| 59 | SYNGR2 | SYNGR2 | 15178 | -0.003 | -0.2667 | No |
| 60 | YWHAE | YWHAE | 15236 | -0.004 | -0.2689 | No |
| 61 | GMDS | GMDS | 15288 | -0.004 | -0.2709 | No |
| 62 | LTBR | LTBR | 15315 | -0.004 | -0.2719 | No |
| 63 | CCND3 | CCND3 | 15462 | -0.006 | -0.2778 | No |
| 64 | TRAPPC10 | TRAPPC10 | 15570 | -0.006 | -0.2820 | No |
| 65 | PHLPP1 | PHLPP1 | 15580 | -0.006 | -0.2823 | No |
| 66 | ME1 | ME1 | 15606 | -0.007 | -0.2831 | No |
| 67 | TRIM24 | TRIM24 | 15878 | -0.009 | -0.2941 | No |
| 68 | TRAF3IP1 | TRAF3IP1 | 16344 | -0.014 | -0.3129 | No |
| 69 | SLC9A7 | SLC9A7 | 16347 | -0.014 | -0.3126 | No |
| 70 | HMBOX1 | HMBOX1 | 16348 | -0.014 | -0.3123 | No |
| 71 | IPPK | IPPK | 16359 | -0.014 | -0.3124 | No |
| 72 | DCAF7 | DCAF7 | 16672 | -0.017 | -0.3248 | No |
| 73 | PTPRU | PTPRU | 16901 | -0.020 | -0.3337 | No |
| 74 | STARD5 | STARD5 | 16917 | -0.020 | -0.3338 | No |
| 75 | PTPRD | PTPRD | 16987 | -0.021 | -0.3362 | No |
| 76 | TCIRG1 | TCIRG1 | 17040 | -0.021 | -0.3378 | No |
| 77 | TCF12 | TCF12 | 17083 | -0.022 | -0.3390 | No |
| 78 | ZFR | ZFR | 17347 | -0.025 | -0.3492 | No |
| 79 | MID2 | MID2 | 18164 | -0.037 | -0.3819 | No |
| 80 | FAM114A1 | FAM114A1 | 18380 | -0.041 | -0.3898 | No |
| 81 | TRAF1 | TRAF1 | 18411 | -0.042 | -0.3900 | No |
| 82 | IQSEC2 | IQSEC2 | 18533 | -0.044 | -0.3939 | No |
| 83 | CBR1 | CBR1 | 18632 | -0.046 | -0.3968 | No |
| 84 | PVR | PVR | 18703 | -0.048 | -0.3985 | No |
| 85 | SMARCD2 | SMARCD2 | 18710 | -0.048 | -0.3975 | No |
| 86 | ABL2 | ABL2 | 18836 | -0.051 | -0.4014 | No |
| 87 | UBE2Z | UBE2Z | 18869 | -0.052 | -0.4015 | No |
| 88 | DNASE1L1 | DNASE1L1 | 18958 | -0.054 | -0.4038 | No |
| 89 | AOX1 | AOX1 | 18974 | -0.055 | -0.4031 | No |
| 90 | CDC42EP1 | CDC42EP1 | 19008 | -0.055 | -0.4031 | No |
| 91 | LLGL2 | LLGL2 | 19126 | -0.059 | -0.4064 | No |
| 92 | STX11 | STX11 | 19243 | -0.063 | -0.4097 | No |
| 93 | IGFBP6 | IGFBP6 | 19469 | -0.071 | -0.4172 | No |
| 94 | ADARB1 | ADARB1 | 19474 | -0.071 | -0.4156 | No |
| 95 | ID4 | ID4 | 19509 | -0.072 | -0.4152 | No |
| 96 | SDC1 | SDC1 | 19569 | -0.074 | -0.4158 | No |
| 97 | ADRB2 | ADRB2 | 19584 | -0.075 | -0.4146 | No |
| 98 | SVIL | SVIL | 19612 | -0.076 | -0.4138 | No |
| 99 | EXPH5 | EXPH5 | 19675 | -0.079 | -0.4144 | No |
| 100 | PLP2 | PLP2 | 19799 | -0.084 | -0.4174 | No |
| 101 | RAB27A | RAB27A | 19809 | -0.084 | -0.4157 | No |
| 102 | CDS1 | CDS1 | 19828 | -0.085 | -0.4144 | No |
| 103 | ASPH | ASPH | 20458 | -0.115 | -0.4375 | No |
| 104 | STAT6 | STAT6 | 20462 | -0.115 | -0.4348 | No |
| 105 | DOCK2 | DOCK2 | 20595 | -0.122 | -0.4372 | No |
| 106 | S100A8 | S100A8 | 20615 | -0.124 | -0.4349 | No |
| 107 | AGPAT2 | AGPAT2 | 21062 | -0.158 | -0.4494 | No |
| 108 | LAMB3 | LAMB3 | 21129 | -0.163 | -0.4481 | No |
| 109 | SH3TC1 | SH3TC1 | 21268 | -0.178 | -0.4494 | No |
| 110 | PLEK2 | PLEK2 | 21279 | -0.180 | -0.4454 | No |
| 111 | IL1R1 | IL1R1 | 21433 | -0.197 | -0.4469 | No |
| 112 | S100A2 | S100A2 | 21627 | -0.215 | -0.4495 | Yes |
| 113 | HTR7 | HTR7 | 21677 | -0.220 | -0.4461 | Yes |
| 114 | FMNL1 | FMNL1 | 21696 | -0.223 | -0.4414 | Yes |
| 115 | SLC10A3 | SLC10A3 | 21813 | -0.241 | -0.4402 | Yes |
| 116 | RIPK4 | RIPK4 | 21834 | -0.244 | -0.4350 | Yes |
| 117 | TMEM40 | TMEM40 | 22312 | -0.323 | -0.4467 | Yes |
| 118 | CHST15 | CHST15 | 22349 | -0.332 | -0.4400 | Yes |
| 119 | TNNC2 | TNNC2 | 22437 | -0.348 | -0.4351 | Yes |
| 120 | PRICKLE3 | PRICKLE3 | 22581 | -0.352 | -0.4323 | Yes |
| 121 | GPRC5A | GPRC5A | 23055 | -0.372 | -0.4426 | Yes |
| 122 | SELPLG | SELPLG | 23120 | -0.387 | -0.4357 | Yes |
| 123 | MSX2 | MSX2 | 23414 | -0.451 | -0.4367 | Yes |
| 124 | PKP3 | PKP3 | 23445 | -0.466 | -0.4264 | Yes |
| 125 | STEAP1 | STEAP1 | 23509 | -0.487 | -0.4170 | Yes |
| 126 | HMGA1 | HMGA1 | 23528 | -0.495 | -0.4056 | Yes |
| 127 | SPINT1 | SPINT1 | 23540 | -0.501 | -0.3937 | Yes |
| 128 | GCNT1 | GCNT1 | 23676 | -0.557 | -0.3855 | Yes |
| 129 | ABLIM3 | ABLIM3 | 23703 | -0.575 | -0.3724 | Yes |
| 130 | TNNT1 | TNNT1 | 23710 | -0.582 | -0.3583 | Yes |
| 131 | COL17A1 | COL17A1 | 23778 | -0.634 | -0.3455 | Yes |
| 132 | BANK1 | BANK1 | 23998 | -0.735 | -0.3364 | Yes |
| 133 | FOSL1 | FOSL1 | 24005 | -0.742 | -0.3183 | Yes |
| 134 | MME | MME | 24089 | -0.848 | -0.3008 | Yes |
| 135 | LAMC2 | LAMC2 | 24233 | -1.143 | -0.2786 | Yes |
| 136 | ALDH3A1 | ALDH3A1 | 24250 | -1.205 | -0.2495 | Yes |
| 137 | LRRC8E | LRRC8E | 24263 | -1.247 | -0.2193 | Yes |
| 138 | MYO1D | MYO1D | 24282 | -1.317 | -0.1875 | Yes |
| 139 | TFAP2C | TFAP2C | 24290 | -1.355 | -0.1544 | Yes |
| 140 | TNFAIP2 | TNFAIP2 | 24295 | -1.370 | -0.1208 | Yes |
| 141 | EVI2A | EVI2A | 24308 | -1.428 | -0.0861 | Yes |
| 142 | PI15 | PI15 | 24353 | -1.762 | -0.0445 | Yes |
| 143 | TBX3 | TBX3 | 24362 | -1.863 | 0.0011 | Yes |