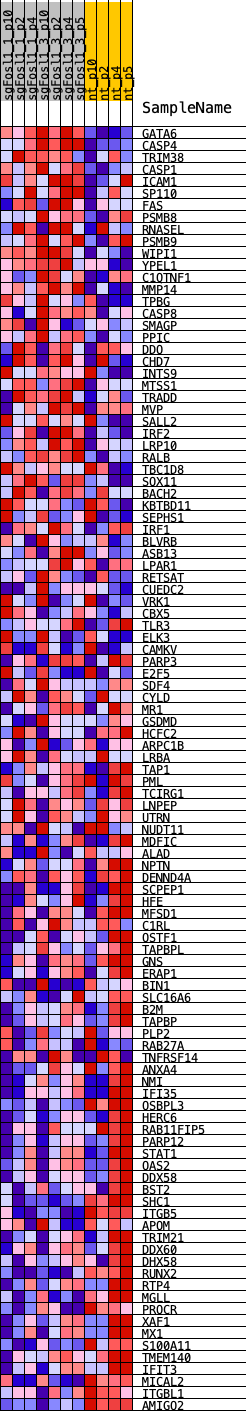
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | SP100_REGULON |
| Enrichment Score (ES) | -0.37118515 |
| Normalized Enrichment Score (NES) | -0.9211689 |
| Nominal p-value | 0.6072961 |
| FDR q-value | 0.6479768 |
| FWER p-Value | 0.992 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GATA6 | GATA6 | 117 | 0.805 | 0.0429 | No |
| 2 | CASP4 | CASP4 | 204 | 0.693 | 0.0804 | No |
| 3 | TRIM38 | TRIM38 | 474 | 0.539 | 0.1012 | No |
| 4 | CASP1 | CASP1 | 480 | 0.536 | 0.1327 | No |
| 5 | ICAM1 | ICAM1 | 511 | 0.517 | 0.1620 | No |
| 6 | SP110 | SP110 | 612 | 0.467 | 0.1856 | No |
| 7 | FAS | FAS | 1384 | 0.309 | 0.1721 | No |
| 8 | PSMB8 | PSMB8 | 1622 | 0.268 | 0.1782 | No |
| 9 | RNASEL | RNASEL | 1770 | 0.253 | 0.1872 | No |
| 10 | PSMB9 | PSMB9 | 2116 | 0.246 | 0.1875 | No |
| 11 | WIPI1 | WIPI1 | 2844 | 0.202 | 0.1695 | No |
| 12 | YPEL1 | YPEL1 | 2919 | 0.193 | 0.1779 | No |
| 13 | C1QTNF1 | C1QTNF1 | 3582 | 0.134 | 0.1586 | No |
| 14 | MMP14 | MMP14 | 4214 | 0.102 | 0.1387 | No |
| 15 | TPBG | TPBG | 4241 | 0.101 | 0.1435 | No |
| 16 | CASP8 | CASP8 | 4344 | 0.096 | 0.1450 | No |
| 17 | SMAGP | SMAGP | 4461 | 0.090 | 0.1456 | No |
| 18 | PPIC | PPIC | 4755 | 0.080 | 0.1382 | No |
| 19 | DDO | DDO | 4979 | 0.073 | 0.1334 | No |
| 20 | CHD7 | CHD7 | 5075 | 0.070 | 0.1336 | No |
| 21 | INTS9 | INTS9 | 5168 | 0.068 | 0.1339 | No |
| 22 | MTSS1 | MTSS1 | 5265 | 0.065 | 0.1338 | No |
| 23 | TRADD | TRADD | 5278 | 0.065 | 0.1371 | No |
| 24 | MVP | MVP | 5446 | 0.061 | 0.1339 | No |
| 25 | SALL2 | SALL2 | 5505 | 0.060 | 0.1351 | No |
| 26 | IRF2 | IRF2 | 5857 | 0.053 | 0.1237 | No |
| 27 | LRP10 | LRP10 | 6021 | 0.050 | 0.1200 | No |
| 28 | RALB | RALB | 6032 | 0.050 | 0.1226 | No |
| 29 | TBC1D8 | TBC1D8 | 6121 | 0.049 | 0.1218 | No |
| 30 | SOX11 | SOX11 | 6421 | 0.044 | 0.1121 | No |
| 31 | BACH2 | BACH2 | 6604 | 0.041 | 0.1070 | No |
| 32 | KBTBD11 | KBTBD11 | 7068 | 0.035 | 0.0900 | No |
| 33 | SEPHS1 | SEPHS1 | 7329 | 0.032 | 0.0812 | No |
| 34 | IRF1 | IRF1 | 7515 | 0.029 | 0.0753 | No |
| 35 | BLVRB | BLVRB | 7872 | 0.025 | 0.0621 | No |
| 36 | ASB13 | ASB13 | 7945 | 0.025 | 0.0606 | No |
| 37 | LPAR1 | LPAR1 | 7985 | 0.024 | 0.0604 | No |
| 38 | RETSAT | RETSAT | 8213 | 0.022 | 0.0523 | No |
| 39 | CUEDC2 | CUEDC2 | 8645 | 0.017 | 0.0356 | No |
| 40 | VRK1 | VRK1 | 8692 | 0.017 | 0.0347 | No |
| 41 | CBX5 | CBX5 | 8837 | 0.016 | 0.0297 | No |
| 42 | TLR3 | TLR3 | 8873 | 0.015 | 0.0292 | No |
| 43 | ELK3 | ELK3 | 9302 | 0.012 | 0.0123 | No |
| 44 | CAMKV | CAMKV | 9866 | 0.007 | -0.0105 | No |
| 45 | PARP3 | PARP3 | 10060 | 0.005 | -0.0182 | No |
| 46 | E2F5 | E2F5 | 10327 | 0.003 | -0.0290 | No |
| 47 | SDF4 | SDF4 | 10358 | 0.002 | -0.0301 | No |
| 48 | CYLD | CYLD | 10624 | 0.000 | -0.0410 | No |
| 49 | MR1 | MR1 | 15125 | -0.003 | -0.2262 | No |
| 50 | GSDMD | GSDMD | 15450 | -0.005 | -0.2392 | No |
| 51 | HCFC2 | HCFC2 | 15525 | -0.006 | -0.2419 | No |
| 52 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.2452 | No |
| 53 | LRBA | LRBA | 15767 | -0.008 | -0.2509 | No |
| 54 | TAP1 | TAP1 | 15960 | -0.010 | -0.2582 | No |
| 55 | PML | PML | 16671 | -0.017 | -0.2864 | No |
| 56 | TCIRG1 | TCIRG1 | 17040 | -0.021 | -0.3003 | No |
| 57 | LNPEP | LNPEP | 17143 | -0.023 | -0.3031 | No |
| 58 | UTRN | UTRN | 17557 | -0.028 | -0.3185 | No |
| 59 | NUDT11 | NUDT11 | 17661 | -0.029 | -0.3210 | No |
| 60 | MDFIC | MDFIC | 17704 | -0.030 | -0.3209 | No |
| 61 | ALAD | ALAD | 17761 | -0.031 | -0.3214 | No |
| 62 | NPTN | NPTN | 18240 | -0.039 | -0.3388 | No |
| 63 | DENND4A | DENND4A | 18290 | -0.040 | -0.3385 | No |
| 64 | SCPEP1 | SCPEP1 | 18472 | -0.043 | -0.3434 | No |
| 65 | HFE | HFE | 18708 | -0.048 | -0.3502 | No |
| 66 | MFSD1 | MFSD1 | 19110 | -0.058 | -0.3633 | Yes |
| 67 | C1RL | C1RL | 19124 | -0.059 | -0.3603 | Yes |
| 68 | OSTF1 | OSTF1 | 19137 | -0.059 | -0.3573 | Yes |
| 69 | TAPBPL | TAPBPL | 19198 | -0.061 | -0.3562 | Yes |
| 70 | GNS | GNS | 19234 | -0.063 | -0.3539 | Yes |
| 71 | ERAP1 | ERAP1 | 19253 | -0.063 | -0.3509 | Yes |
| 72 | BIN1 | BIN1 | 19395 | -0.068 | -0.3526 | Yes |
| 73 | SLC16A6 | SLC16A6 | 19560 | -0.074 | -0.3550 | Yes |
| 74 | B2M | B2M | 19601 | -0.075 | -0.3522 | Yes |
| 75 | TAPBP | TAPBP | 19699 | -0.079 | -0.3515 | Yes |
| 76 | PLP2 | PLP2 | 19799 | -0.084 | -0.3506 | Yes |
| 77 | RAB27A | RAB27A | 19809 | -0.084 | -0.3460 | Yes |
| 78 | TNFRSF14 | TNFRSF14 | 20165 | -0.102 | -0.3546 | Yes |
| 79 | ANXA4 | ANXA4 | 20186 | -0.103 | -0.3493 | Yes |
| 80 | NMI | NMI | 20390 | -0.112 | -0.3511 | Yes |
| 81 | IFI35 | IFI35 | 20825 | -0.135 | -0.3610 | Yes |
| 82 | OSBPL3 | OSBPL3 | 21074 | -0.159 | -0.3618 | Yes |
| 83 | HERC6 | HERC6 | 21103 | -0.161 | -0.3534 | Yes |
| 84 | RAB11FIP5 | RAB11FIP5 | 21132 | -0.163 | -0.3449 | Yes |
| 85 | PARP12 | PARP12 | 21383 | -0.192 | -0.3438 | Yes |
| 86 | STAT1 | STAT1 | 21387 | -0.192 | -0.3326 | Yes |
| 87 | OAS2 | OAS2 | 21388 | -0.192 | -0.3212 | Yes |
| 88 | DDX58 | DDX58 | 21453 | -0.199 | -0.3121 | Yes |
| 89 | BST2 | BST2 | 21635 | -0.216 | -0.3067 | Yes |
| 90 | SHC1 | SHC1 | 21741 | -0.230 | -0.2974 | Yes |
| 91 | ITGB5 | ITGB5 | 21764 | -0.233 | -0.2845 | Yes |
| 92 | APOM | APOM | 21932 | -0.258 | -0.2761 | Yes |
| 93 | TRIM21 | TRIM21 | 21956 | -0.263 | -0.2615 | Yes |
| 94 | DDX60 | DDX60 | 22034 | -0.275 | -0.2484 | Yes |
| 95 | DHX58 | DHX58 | 22071 | -0.281 | -0.2332 | Yes |
| 96 | RUNX2 | RUNX2 | 22102 | -0.286 | -0.2175 | Yes |
| 97 | RTP4 | RTP4 | 22220 | -0.305 | -0.2043 | Yes |
| 98 | MGLL | MGLL | 22225 | -0.306 | -0.1864 | Yes |
| 99 | PROCR | PROCR | 22311 | -0.323 | -0.1707 | Yes |
| 100 | XAF1 | XAF1 | 22417 | -0.344 | -0.1547 | Yes |
| 101 | MX1 | MX1 | 22631 | -0.352 | -0.1426 | Yes |
| 102 | S100A11 | S100A11 | 23510 | -0.487 | -0.1499 | Yes |
| 103 | TMEM140 | TMEM140 | 23515 | -0.489 | -0.1211 | Yes |
| 104 | IFIT3 | IFIT3 | 23546 | -0.503 | -0.0926 | Yes |
| 105 | MICAL2 | MICAL2 | 23614 | -0.532 | -0.0639 | Yes |
| 106 | ITGBL1 | ITGBL1 | 23932 | -0.682 | -0.0366 | Yes |
| 107 | AMIGO2 | AMIGO2 | 24139 | -0.934 | 0.0103 | Yes |