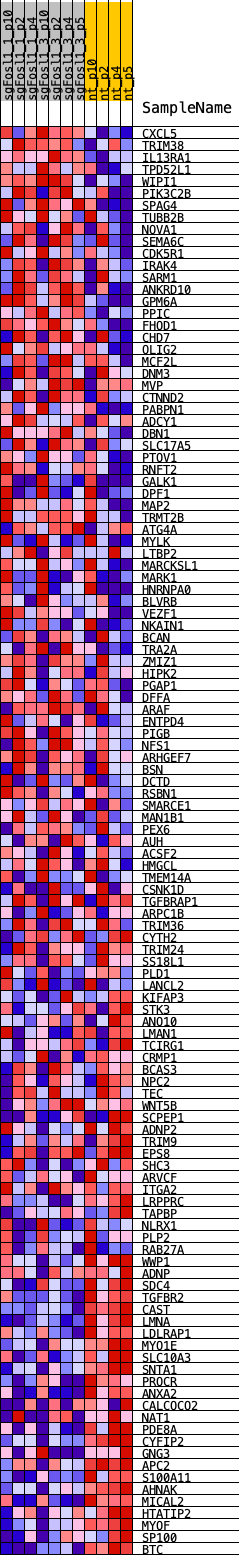
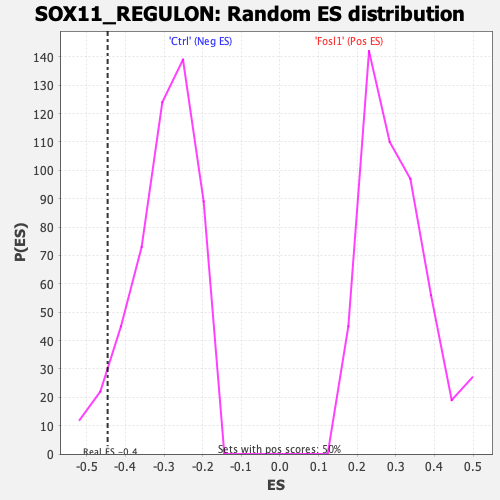
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | SOX11_REGULON |
| Enrichment Score (ES) | -0.4462616 |
| Normalized Enrichment Score (NES) | -1.4732685 |
| Nominal p-value | 0.061507937 |
| FDR q-value | 0.086715296 |
| FWER p-Value | 0.269 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CXCL5 | CXCL5 | 44 | 1.039 | 0.0655 | No |
| 2 | TRIM38 | TRIM38 | 474 | 0.539 | 0.0827 | No |
| 3 | IL13RA1 | IL13RA1 | 649 | 0.452 | 0.1048 | No |
| 4 | TPD52L1 | TPD52L1 | 2689 | 0.223 | 0.0353 | No |
| 5 | WIPI1 | WIPI1 | 2844 | 0.202 | 0.0420 | No |
| 6 | PIK3C2B | PIK3C2B | 3165 | 0.169 | 0.0398 | No |
| 7 | SPAG4 | SPAG4 | 3200 | 0.167 | 0.0492 | No |
| 8 | TUBB2B | TUBB2B | 3621 | 0.132 | 0.0404 | No |
| 9 | NOVA1 | NOVA1 | 3979 | 0.112 | 0.0330 | No |
| 10 | SEMA6C | SEMA6C | 4059 | 0.109 | 0.0367 | No |
| 11 | CDK5R1 | CDK5R1 | 4069 | 0.108 | 0.0434 | No |
| 12 | IRAK4 | IRAK4 | 4188 | 0.103 | 0.0452 | No |
| 13 | SARM1 | SARM1 | 4279 | 0.099 | 0.0479 | No |
| 14 | ANKRD10 | ANKRD10 | 4374 | 0.094 | 0.0502 | No |
| 15 | GPM6A | GPM6A | 4657 | 0.082 | 0.0439 | No |
| 16 | PPIC | PPIC | 4755 | 0.080 | 0.0450 | No |
| 17 | FHOD1 | FHOD1 | 4903 | 0.075 | 0.0438 | No |
| 18 | CHD7 | CHD7 | 5075 | 0.070 | 0.0414 | No |
| 19 | OLIG2 | OLIG2 | 5125 | 0.069 | 0.0438 | No |
| 20 | MCF2L | MCF2L | 5192 | 0.067 | 0.0454 | No |
| 21 | DNM3 | DNM3 | 5295 | 0.065 | 0.0454 | No |
| 22 | MVP | MVP | 5446 | 0.061 | 0.0432 | No |
| 23 | CTNND2 | CTNND2 | 5701 | 0.056 | 0.0364 | No |
| 24 | PABPN1 | PABPN1 | 5702 | 0.056 | 0.0400 | No |
| 25 | ADCY1 | ADCY1 | 5752 | 0.055 | 0.0416 | No |
| 26 | DBN1 | DBN1 | 5806 | 0.054 | 0.0429 | No |
| 27 | SLC17A5 | SLC17A5 | 5958 | 0.051 | 0.0400 | No |
| 28 | PTOV1 | PTOV1 | 6003 | 0.050 | 0.0414 | No |
| 29 | RNFT2 | RNFT2 | 6045 | 0.050 | 0.0430 | No |
| 30 | GALK1 | GALK1 | 6253 | 0.046 | 0.0374 | No |
| 31 | DPF1 | DPF1 | 6396 | 0.044 | 0.0344 | No |
| 32 | MAP2 | MAP2 | 6412 | 0.044 | 0.0366 | No |
| 33 | TRMT2B | TRMT2B | 6430 | 0.043 | 0.0387 | No |
| 34 | ATG4A | ATG4A | 6691 | 0.040 | 0.0306 | No |
| 35 | MYLK | MYLK | 6812 | 0.038 | 0.0281 | No |
| 36 | LTBP2 | LTBP2 | 6925 | 0.036 | 0.0259 | No |
| 37 | MARCKSL1 | MARCKSL1 | 7231 | 0.033 | 0.0154 | No |
| 38 | MARK1 | MARK1 | 7288 | 0.032 | 0.0152 | No |
| 39 | HNRNPA0 | HNRNPA0 | 7844 | 0.026 | -0.0060 | No |
| 40 | BLVRB | BLVRB | 7872 | 0.025 | -0.0055 | No |
| 41 | VEZF1 | VEZF1 | 7877 | 0.025 | -0.0040 | No |
| 42 | NKAIN1 | NKAIN1 | 7907 | 0.025 | -0.0036 | No |
| 43 | BCAN | BCAN | 7937 | 0.025 | -0.0032 | No |
| 44 | TRA2A | TRA2A | 7939 | 0.025 | -0.0016 | No |
| 45 | ZMIZ1 | ZMIZ1 | 8257 | 0.021 | -0.0133 | No |
| 46 | HIPK2 | HIPK2 | 8360 | 0.020 | -0.0162 | No |
| 47 | PGAP1 | PGAP1 | 8499 | 0.019 | -0.0207 | No |
| 48 | DFFA | DFFA | 8561 | 0.018 | -0.0220 | No |
| 49 | ARAF | ARAF | 8828 | 0.016 | -0.0320 | No |
| 50 | ENTPD4 | ENTPD4 | 8921 | 0.015 | -0.0348 | No |
| 51 | PIGB | PIGB | 8971 | 0.015 | -0.0359 | No |
| 52 | NFS1 | NFS1 | 8978 | 0.014 | -0.0352 | No |
| 53 | ARHGEF7 | ARHGEF7 | 9004 | 0.014 | -0.0353 | No |
| 54 | BSN | BSN | 9028 | 0.014 | -0.0353 | No |
| 55 | DCTD | DCTD | 9383 | 0.011 | -0.0492 | No |
| 56 | RSBN1 | RSBN1 | 9520 | 0.010 | -0.0542 | No |
| 57 | SMARCE1 | SMARCE1 | 9806 | 0.007 | -0.0655 | No |
| 58 | MAN1B1 | MAN1B1 | 9940 | 0.006 | -0.0706 | No |
| 59 | PEX6 | PEX6 | 9984 | 0.005 | -0.0720 | No |
| 60 | AUH | AUH | 10197 | 0.004 | -0.0805 | No |
| 61 | ACSF2 | ACSF2 | 10527 | 0.001 | -0.0940 | No |
| 62 | HMGCL | HMGCL | 10601 | 0.000 | -0.0970 | No |
| 63 | TMEM14A | TMEM14A | 14921 | -0.001 | -0.2749 | No |
| 64 | CSNK1D | CSNK1D | 15475 | -0.006 | -0.2973 | No |
| 65 | TGFBRAP1 | TGFBRAP1 | 15555 | -0.006 | -0.3002 | No |
| 66 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.3022 | No |
| 67 | TRIM36 | TRIM36 | 15631 | -0.007 | -0.3023 | No |
| 68 | CYTH2 | CYTH2 | 15704 | -0.008 | -0.3048 | No |
| 69 | TRIM24 | TRIM24 | 15878 | -0.009 | -0.3113 | No |
| 70 | SS18L1 | SS18L1 | 16024 | -0.011 | -0.3166 | No |
| 71 | PLD1 | PLD1 | 16063 | -0.011 | -0.3174 | No |
| 72 | LANCL2 | LANCL2 | 16095 | -0.012 | -0.3179 | No |
| 73 | KIFAP3 | KIFAP3 | 16252 | -0.013 | -0.3235 | No |
| 74 | STK3 | STK3 | 16608 | -0.017 | -0.3371 | No |
| 75 | ANO10 | ANO10 | 16609 | -0.017 | -0.3360 | No |
| 76 | LMAN1 | LMAN1 | 16911 | -0.020 | -0.3471 | No |
| 77 | TCIRG1 | TCIRG1 | 17040 | -0.021 | -0.3510 | No |
| 78 | CRMP1 | CRMP1 | 17283 | -0.025 | -0.3594 | No |
| 79 | BCAS3 | BCAS3 | 17561 | -0.028 | -0.3690 | No |
| 80 | NPC2 | NPC2 | 17773 | -0.031 | -0.3757 | No |
| 81 | TEC | TEC | 17977 | -0.034 | -0.3818 | No |
| 82 | WNT5B | WNT5B | 18104 | -0.036 | -0.3847 | No |
| 83 | SCPEP1 | SCPEP1 | 18472 | -0.043 | -0.3970 | No |
| 84 | ADNP2 | ADNP2 | 18696 | -0.048 | -0.4031 | No |
| 85 | TRIM9 | TRIM9 | 18789 | -0.050 | -0.4036 | No |
| 86 | EPS8 | EPS8 | 18901 | -0.053 | -0.4048 | No |
| 87 | SHC3 | SHC3 | 19146 | -0.059 | -0.4110 | No |
| 88 | ARVCF | ARVCF | 19161 | -0.060 | -0.4077 | No |
| 89 | ITGA2 | ITGA2 | 19296 | -0.065 | -0.4090 | No |
| 90 | LRPPRC | LRPPRC | 19514 | -0.072 | -0.4133 | No |
| 91 | TAPBP | TAPBP | 19699 | -0.079 | -0.4157 | No |
| 92 | NLRX1 | NLRX1 | 19786 | -0.083 | -0.4139 | No |
| 93 | PLP2 | PLP2 | 19799 | -0.084 | -0.4089 | No |
| 94 | RAB27A | RAB27A | 19809 | -0.084 | -0.4038 | No |
| 95 | WWP1 | WWP1 | 19861 | -0.087 | -0.4003 | No |
| 96 | ADNP | ADNP | 20096 | -0.098 | -0.4035 | No |
| 97 | SDC4 | SDC4 | 20885 | -0.139 | -0.4270 | No |
| 98 | TGFBR2 | TGFBR2 | 21354 | -0.188 | -0.4341 | Yes |
| 99 | CAST | CAST | 21363 | -0.190 | -0.4221 | Yes |
| 100 | LMNA | LMNA | 21572 | -0.210 | -0.4171 | Yes |
| 101 | LDLRAP1 | LDLRAP1 | 21687 | -0.221 | -0.4074 | Yes |
| 102 | MYO1E | MYO1E | 21801 | -0.239 | -0.3966 | Yes |
| 103 | SLC10A3 | SLC10A3 | 21813 | -0.241 | -0.3815 | Yes |
| 104 | SNTA1 | SNTA1 | 22055 | -0.278 | -0.3734 | Yes |
| 105 | PROCR | PROCR | 22311 | -0.323 | -0.3630 | Yes |
| 106 | ANXA2 | ANXA2 | 22374 | -0.334 | -0.3439 | Yes |
| 107 | CALCOCO2 | CALCOCO2 | 22981 | -0.359 | -0.3456 | Yes |
| 108 | NAT1 | NAT1 | 22995 | -0.361 | -0.3227 | Yes |
| 109 | PDE8A | PDE8A | 23122 | -0.389 | -0.3027 | Yes |
| 110 | CYFIP2 | CYFIP2 | 23124 | -0.389 | -0.2776 | Yes |
| 111 | GNG3 | GNG3 | 23454 | -0.469 | -0.2607 | Yes |
| 112 | APC2 | APC2 | 23483 | -0.476 | -0.2311 | Yes |
| 113 | S100A11 | S100A11 | 23510 | -0.487 | -0.2006 | Yes |
| 114 | AHNAK | AHNAK | 23609 | -0.531 | -0.1702 | Yes |
| 115 | MICAL2 | MICAL2 | 23614 | -0.532 | -0.1359 | Yes |
| 116 | HTATIP2 | HTATIP2 | 23654 | -0.546 | -0.1022 | Yes |
| 117 | MYOF | MYOF | 23756 | -0.619 | -0.0662 | Yes |
| 118 | SP100 | SP100 | 23768 | -0.624 | -0.0263 | Yes |
| 119 | BTC | BTC | 24053 | -0.799 | 0.0138 | Yes |