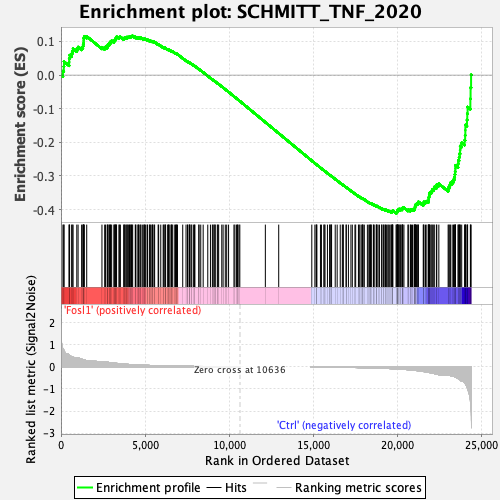
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | SCHMITT_TNF_2020 |
| Enrichment Score (ES) | -0.4104595 |
| Normalized Enrichment Score (NES) | -1.3235664 |
| Nominal p-value | 0.07231405 |
| FDR q-value | 0.14713097 |
| FWER p-Value | 0.628 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PRR16 | PRR16 | 116 | 0.807 | 0.0119 | No |
| 2 | BCL3 | BCL3 | 167 | 0.756 | 0.0255 | No |
| 3 | STAP2 | STAP2 | 178 | 0.729 | 0.0402 | No |
| 4 | VDR | VDR | 484 | 0.533 | 0.0386 | No |
| 5 | KCNN4 | KCNN4 | 487 | 0.528 | 0.0495 | No |
| 6 | ICAM1 | ICAM1 | 511 | 0.517 | 0.0593 | No |
| 7 | KLHL6 | KLHL6 | 633 | 0.462 | 0.0638 | No |
| 8 | LIF | LIF | 668 | 0.445 | 0.0717 | No |
| 9 | DRAM1 | DRAM1 | 718 | 0.434 | 0.0787 | No |
| 10 | NFKBIE | NFKBIE | 939 | 0.413 | 0.0781 | No |
| 11 | HAS2 | HAS2 | 1015 | 0.408 | 0.0835 | No |
| 12 | ENPP2 | ENPP2 | 1225 | 0.348 | 0.0820 | No |
| 13 | VWA1 | VWA1 | 1299 | 0.329 | 0.0858 | No |
| 14 | OSMR | OSMR | 1318 | 0.324 | 0.0918 | No |
| 15 | TMEM151B | TMEM151B | 1344 | 0.318 | 0.0973 | No |
| 16 | BATF3 | BATF3 | 1347 | 0.316 | 0.1038 | No |
| 17 | MYO5B | MYO5B | 1349 | 0.316 | 0.1103 | No |
| 18 | ANO9 | ANO9 | 1383 | 0.309 | 0.1154 | No |
| 19 | SOCS3 | SOCS3 | 1528 | 0.284 | 0.1153 | No |
| 20 | JUNB | JUNB | 2433 | 0.243 | 0.0828 | No |
| 21 | NFE2L3 | NFE2L3 | 2596 | 0.236 | 0.0810 | No |
| 22 | KREMEN2 | KREMEN2 | 2640 | 0.231 | 0.0840 | No |
| 23 | LAPTM5 | LAPTM5 | 2742 | 0.215 | 0.0842 | No |
| 24 | TNFAIP3 | TNFAIP3 | 2772 | 0.211 | 0.0874 | No |
| 25 | IRAK2 | IRAK2 | 2789 | 0.209 | 0.0911 | No |
| 26 | PLAU | PLAU | 2866 | 0.199 | 0.0921 | No |
| 27 | APOL6 | APOL6 | 2872 | 0.198 | 0.0960 | No |
| 28 | FAM174B | FAM174B | 2941 | 0.191 | 0.0971 | No |
| 29 | NFKBIZ | NFKBIZ | 2952 | 0.189 | 0.1006 | No |
| 30 | PLLP | PLLP | 3009 | 0.184 | 0.1021 | No |
| 31 | THY1 | THY1 | 3146 | 0.171 | 0.1000 | No |
| 32 | NR4A1 | NR4A1 | 3163 | 0.170 | 0.1029 | No |
| 33 | ALPK1 | ALPK1 | 3199 | 0.167 | 0.1049 | No |
| 34 | DPP4 | DPP4 | 3244 | 0.163 | 0.1065 | No |
| 35 | WIPF3 | WIPF3 | 3256 | 0.162 | 0.1094 | No |
| 36 | RELB | RELB | 3268 | 0.161 | 0.1122 | No |
| 37 | EBI3 | EBI3 | 3301 | 0.159 | 0.1142 | No |
| 38 | PTP4A3 | PTP4A3 | 3444 | 0.145 | 0.1113 | No |
| 39 | ZFP36 | ZFP36 | 3467 | 0.143 | 0.1134 | No |
| 40 | TIMP1 | TIMP1 | 3528 | 0.139 | 0.1138 | No |
| 41 | NFKB2 | NFKB2 | 3730 | 0.126 | 0.1081 | No |
| 42 | COL9A1 | COL9A1 | 3745 | 0.124 | 0.1101 | No |
| 43 | ZDHHC14 | ZDHHC14 | 3790 | 0.121 | 0.1107 | No |
| 44 | ADAMTS1 | ADAMTS1 | 3813 | 0.120 | 0.1123 | No |
| 45 | CABLES1 | CABLES1 | 3913 | 0.115 | 0.1106 | No |
| 46 | DUSP5 | DUSP5 | 3914 | 0.115 | 0.1130 | No |
| 47 | ROBO1 | ROBO1 | 3970 | 0.112 | 0.1130 | No |
| 48 | TIFA | TIFA | 4022 | 0.111 | 0.1132 | No |
| 49 | PRKAR1B | PRKAR1B | 4072 | 0.108 | 0.1134 | No |
| 50 | LIFR | LIFR | 4103 | 0.106 | 0.1144 | No |
| 51 | RRAS | RRAS | 4145 | 0.105 | 0.1149 | No |
| 52 | SLC16A3 | SLC16A3 | 4187 | 0.103 | 0.1153 | No |
| 53 | PLEKHF1 | PLEKHF1 | 4218 | 0.102 | 0.1162 | No |
| 54 | IL27RA | IL27RA | 4258 | 0.100 | 0.1166 | No |
| 55 | GABRB3 | GABRB3 | 4414 | 0.093 | 0.1121 | No |
| 56 | SEMA3B | SEMA3B | 4424 | 0.092 | 0.1136 | No |
| 57 | C4A | C4A | 4520 | 0.088 | 0.1115 | No |
| 58 | BIRC2 | BIRC2 | 4593 | 0.085 | 0.1103 | No |
| 59 | SORBS1 | SORBS1 | 4601 | 0.084 | 0.1117 | No |
| 60 | CNTNAP1 | CNTNAP1 | 4644 | 0.083 | 0.1117 | No |
| 61 | GRB10 | GRB10 | 4738 | 0.080 | 0.1095 | No |
| 62 | STAT3 | STAT3 | 4744 | 0.080 | 0.1110 | No |
| 63 | RASEF | RASEF | 4853 | 0.076 | 0.1081 | No |
| 64 | NRCAM | NRCAM | 4924 | 0.075 | 0.1067 | No |
| 65 | PIK3CD | PIK3CD | 4946 | 0.074 | 0.1074 | No |
| 66 | IFNGR2 | IFNGR2 | 4973 | 0.073 | 0.1078 | No |
| 67 | NFKBIA | NFKBIA | 5028 | 0.072 | 0.1071 | No |
| 68 | CITED1 | CITED1 | 5116 | 0.069 | 0.1049 | No |
| 69 | MPZ | MPZ | 5142 | 0.069 | 0.1053 | No |
| 70 | LRRC4B | LRRC4B | 5250 | 0.066 | 0.1022 | No |
| 71 | F3 | F3 | 5284 | 0.065 | 0.1022 | No |
| 72 | GAP43 | GAP43 | 5306 | 0.064 | 0.1026 | No |
| 73 | APBA1 | APBA1 | 5397 | 0.062 | 0.1002 | No |
| 74 | ARHGEF40 | ARHGEF40 | 5438 | 0.062 | 0.0998 | No |
| 75 | KLHL5 | KLHL5 | 5452 | 0.061 | 0.1005 | No |
| 76 | ALPL | ALPL | 5529 | 0.059 | 0.0986 | No |
| 77 | RAB13 | RAB13 | 5563 | 0.059 | 0.0985 | No |
| 78 | RNF19A | RNF19A | 5772 | 0.054 | 0.0910 | No |
| 79 | ETV6 | ETV6 | 5802 | 0.054 | 0.0909 | No |
| 80 | LIMD2 | LIMD2 | 5927 | 0.052 | 0.0868 | No |
| 81 | EGR2 | EGR2 | 6070 | 0.049 | 0.0819 | No |
| 82 | PDGFA | PDGFA | 6104 | 0.049 | 0.0816 | No |
| 83 | BCL6 | BCL6 | 6184 | 0.048 | 0.0793 | No |
| 84 | CEBPB | CEBPB | 6185 | 0.048 | 0.0803 | No |
| 85 | AGT | AGT | 6195 | 0.047 | 0.0809 | No |
| 86 | PLEKHA4 | PLEKHA4 | 6320 | 0.045 | 0.0767 | No |
| 87 | LOXL2 | LOXL2 | 6389 | 0.044 | 0.0748 | No |
| 88 | TNIP2 | TNIP2 | 6402 | 0.044 | 0.0752 | No |
| 89 | ZIK1 | ZIK1 | 6426 | 0.043 | 0.0751 | No |
| 90 | SLC16A9 | SLC16A9 | 6530 | 0.042 | 0.0717 | No |
| 91 | IER2 | IER2 | 6568 | 0.041 | 0.0711 | No |
| 92 | CDH4 | CDH4 | 6583 | 0.041 | 0.0713 | No |
| 93 | TAF13 | TAF13 | 6638 | 0.040 | 0.0699 | No |
| 94 | TNFRSF1A | TNFRSF1A | 6759 | 0.039 | 0.0658 | No |
| 95 | PCDH1 | PCDH1 | 6804 | 0.038 | 0.0647 | No |
| 96 | CXCL10 | CXCL10 | 6851 | 0.037 | 0.0636 | No |
| 97 | ACSL6 | ACSL6 | 6868 | 0.037 | 0.0637 | No |
| 98 | ORAI1 | ORAI1 | 6920 | 0.036 | 0.0623 | No |
| 99 | TBC1D10A | TBC1D10A | 7232 | 0.033 | 0.0501 | No |
| 100 | NPNT | NPNT | 7429 | 0.030 | 0.0426 | No |
| 101 | IRF1 | IRF1 | 7515 | 0.029 | 0.0397 | No |
| 102 | FJX1 | FJX1 | 7533 | 0.029 | 0.0396 | No |
| 103 | PLA2G4C | PLA2G4C | 7624 | 0.028 | 0.0365 | No |
| 104 | TSC22D4 | TSC22D4 | 7627 | 0.028 | 0.0369 | No |
| 105 | SYNGR3 | SYNGR3 | 7655 | 0.028 | 0.0364 | No |
| 106 | RBM3 | RBM3 | 7727 | 0.027 | 0.0340 | No |
| 107 | PNKD | PNKD | 7771 | 0.027 | 0.0328 | No |
| 108 | ARRDC4 | ARRDC4 | 7879 | 0.025 | 0.0289 | No |
| 109 | CSF1 | CSF1 | 7934 | 0.025 | 0.0271 | No |
| 110 | ZSWIM4 | ZSWIM4 | 7955 | 0.024 | 0.0268 | No |
| 111 | SPRED3 | SPRED3 | 8190 | 0.022 | 0.0176 | No |
| 112 | KLF10 | KLF10 | 8197 | 0.022 | 0.0178 | No |
| 113 | RNF145 | RNF145 | 8303 | 0.021 | 0.0138 | No |
| 114 | TGFB1 | TGFB1 | 8447 | 0.019 | 0.0083 | No |
| 115 | RGS3 | RGS3 | 8720 | 0.017 | -0.0026 | No |
| 116 | S100A10 | S100A10 | 8879 | 0.015 | -0.0089 | No |
| 117 | NFKB1 | NFKB1 | 8993 | 0.014 | -0.0133 | No |
| 118 | ARNTL2 | ARNTL2 | 9035 | 0.014 | -0.0147 | No |
| 119 | NDUFB8 | NDUFB8 | 9097 | 0.013 | -0.0169 | No |
| 120 | RPGR | RPGR | 9152 | 0.013 | -0.0189 | No |
| 121 | ITGAV | ITGAV | 9156 | 0.013 | -0.0188 | No |
| 122 | PGBD5 | PGBD5 | 9287 | 0.012 | -0.0239 | No |
| 123 | EFTUD2 | EFTUD2 | 9320 | 0.011 | -0.0250 | No |
| 124 | TAGLN2 | TAGLN2 | 9367 | 0.011 | -0.0267 | No |
| 125 | ANKRD33B | ANKRD33B | 9552 | 0.009 | -0.0341 | No |
| 126 | ADAMTS4 | ADAMTS4 | 9630 | 0.009 | -0.0371 | No |
| 127 | ATCAY | ATCAY | 9761 | 0.007 | -0.0424 | No |
| 128 | IRF3 | IRF3 | 9844 | 0.007 | -0.0456 | No |
| 129 | CDKAL1 | CDKAL1 | 9956 | 0.006 | -0.0501 | No |
| 130 | ANXA1 | ANXA1 | 10276 | 0.003 | -0.0633 | No |
| 131 | PMP22 | PMP22 | 10332 | 0.003 | -0.0655 | No |
| 132 | NTN1 | NTN1 | 10431 | 0.002 | -0.0696 | No |
| 133 | IFNGR1 | IFNGR1 | 10479 | 0.001 | -0.0715 | No |
| 134 | PTX3 | PTX3 | 10494 | 0.001 | -0.0720 | No |
| 135 | MGAT3 | MGAT3 | 10570 | 0.001 | -0.0751 | No |
| 136 | CYLD | CYLD | 10624 | 0.000 | -0.0773 | No |
| 137 | CERS1 | CERS1 | 12141 | 0.000 | -0.1402 | No |
| 138 | GTSF1 | GTSF1 | 12935 | 0.000 | -0.1731 | No |
| 139 | SOD2 | SOD2 | 14901 | -0.001 | -0.2547 | No |
| 140 | PLCD3 | PLCD3 | 15085 | -0.002 | -0.2622 | No |
| 141 | HIVEP1 | HIVEP1 | 15161 | -0.003 | -0.2653 | No |
| 142 | MAP3K11 | MAP3K11 | 15189 | -0.003 | -0.2663 | No |
| 143 | NCOA7 | NCOA7 | 15197 | -0.003 | -0.2665 | No |
| 144 | CSRNP1 | CSRNP1 | 15415 | -0.005 | -0.2754 | No |
| 145 | TMEM158 | TMEM158 | 15473 | -0.006 | -0.2777 | No |
| 146 | ELL2 | ELL2 | 15622 | -0.007 | -0.2837 | No |
| 147 | BMP2 | BMP2 | 15635 | -0.007 | -0.2840 | No |
| 148 | ARHGEF4 | ARHGEF4 | 15686 | -0.008 | -0.2859 | No |
| 149 | SERPINE1 | SERPINE1 | 15830 | -0.009 | -0.2917 | No |
| 150 | ADAM19 | ADAM19 | 15961 | -0.010 | -0.2969 | No |
| 151 | GCLM | GCLM | 15976 | -0.010 | -0.2972 | No |
| 152 | MGAT4B | MGAT4B | 16035 | -0.011 | -0.2994 | No |
| 153 | ARL6IP5 | ARL6IP5 | 16052 | -0.011 | -0.2998 | No |
| 154 | RIPK1 | RIPK1 | 16076 | -0.011 | -0.3006 | No |
| 155 | IL6ST | IL6ST | 16292 | -0.014 | -0.3092 | No |
| 156 | ADAMTS9 | ADAMTS9 | 16408 | -0.015 | -0.3137 | No |
| 157 | SEMA4B | SEMA4B | 16590 | -0.016 | -0.3208 | No |
| 158 | SLC35F6 | SLC35F6 | 16714 | -0.018 | -0.3256 | No |
| 159 | EMP3 | EMP3 | 16762 | -0.018 | -0.3272 | No |
| 160 | OSBPL5 | OSBPL5 | 16764 | -0.018 | -0.3268 | No |
| 161 | RDH10 | RDH10 | 16938 | -0.020 | -0.3336 | No |
| 162 | SPARCL1 | SPARCL1 | 16951 | -0.021 | -0.3336 | No |
| 163 | CD63 | CD63 | 17076 | -0.022 | -0.3383 | No |
| 164 | HMOX1 | HMOX1 | 17226 | -0.024 | -0.3440 | No |
| 165 | MATN2 | MATN2 | 17308 | -0.025 | -0.3469 | No |
| 166 | VMP1 | VMP1 | 17464 | -0.027 | -0.3527 | No |
| 167 | RAB35 | RAB35 | 17508 | -0.027 | -0.3540 | No |
| 168 | GRWD1 | GRWD1 | 17672 | -0.030 | -0.3601 | No |
| 169 | SGK1 | SGK1 | 17712 | -0.030 | -0.3611 | No |
| 170 | TRIM8 | TRIM8 | 17765 | -0.031 | -0.3626 | No |
| 171 | INSR | INSR | 17870 | -0.032 | -0.3663 | No |
| 172 | RHPN2 | RHPN2 | 17908 | -0.033 | -0.3671 | No |
| 173 | TMEM173 | TMEM173 | 17927 | -0.033 | -0.3672 | No |
| 174 | SFRP1 | SFRP1 | 17944 | -0.034 | -0.3671 | No |
| 175 | WWTR1 | WWTR1 | 18020 | -0.035 | -0.3695 | No |
| 176 | CREB3 | CREB3 | 18203 | -0.038 | -0.3763 | No |
| 177 | OSCP1 | OSCP1 | 18289 | -0.040 | -0.3790 | No |
| 178 | SNX10 | SNX10 | 18335 | -0.040 | -0.3800 | No |
| 179 | PSMD5 | PSMD5 | 18387 | -0.041 | -0.3813 | No |
| 180 | TRAF1 | TRAF1 | 18411 | -0.042 | -0.3814 | No |
| 181 | IFITM1 | IFITM1 | 18435 | -0.042 | -0.3814 | No |
| 182 | PLAT | PLAT | 18557 | -0.045 | -0.3855 | No |
| 183 | SMOC2 | SMOC2 | 18570 | -0.045 | -0.3851 | No |
| 184 | CARS | CARS | 18644 | -0.047 | -0.3872 | No |
| 185 | PDGFRL | PDGFRL | 18743 | -0.049 | -0.3902 | No |
| 186 | DUSP6 | DUSP6 | 18759 | -0.049 | -0.3898 | No |
| 187 | PRICKLE1 | PRICKLE1 | 18766 | -0.050 | -0.3890 | No |
| 188 | UGCG | UGCG | 18841 | -0.051 | -0.3910 | No |
| 189 | LGALS1 | LGALS1 | 18926 | -0.053 | -0.3934 | No |
| 190 | OPTN | OPTN | 19056 | -0.057 | -0.3976 | No |
| 191 | PEPD | PEPD | 19059 | -0.057 | -0.3965 | No |
| 192 | NUAK2 | NUAK2 | 19150 | -0.060 | -0.3990 | No |
| 193 | FOSL2 | FOSL2 | 19180 | -0.061 | -0.3989 | No |
| 194 | ERAP1 | ERAP1 | 19253 | -0.063 | -0.4006 | No |
| 195 | EMILIN1 | EMILIN1 | 19277 | -0.064 | -0.4002 | No |
| 196 | RAB31 | RAB31 | 19332 | -0.066 | -0.4011 | No |
| 197 | VAX1 | VAX1 | 19421 | -0.069 | -0.4033 | No |
| 198 | KLF5 | KLF5 | 19442 | -0.070 | -0.4027 | No |
| 199 | MITD1 | MITD1 | 19528 | -0.073 | -0.4047 | No |
| 200 | B2M | B2M | 19601 | -0.075 | -0.4061 | No |
| 201 | NAMPT | NAMPT | 19644 | -0.077 | -0.4063 | No |
| 202 | RELL1 | RELL1 | 19671 | -0.078 | -0.4057 | No |
| 203 | TAPBP | TAPBP | 19699 | -0.079 | -0.4052 | No |
| 204 | CDKN1A | CDKN1A | 19702 | -0.080 | -0.4036 | No |
| 205 | PXDN | PXDN | 19712 | -0.080 | -0.4024 | No |
| 206 | CRABP2 | CRABP2 | 19907 | -0.089 | -0.4086 | No |
| 207 | CD44 | CD44 | 19954 | -0.091 | -0.4086 | Yes |
| 208 | TNC | TNC | 19967 | -0.092 | -0.4072 | Yes |
| 209 | LPGAT1 | LPGAT1 | 19980 | -0.093 | -0.4057 | Yes |
| 210 | SERTAD1 | SERTAD1 | 19991 | -0.093 | -0.4042 | Yes |
| 211 | WARS | WARS | 19999 | -0.094 | -0.4026 | Yes |
| 212 | PPP1R18 | PPP1R18 | 20006 | -0.094 | -0.4009 | Yes |
| 213 | PODXL | PODXL | 20071 | -0.097 | -0.4015 | Yes |
| 214 | SQSTM1 | SQSTM1 | 20077 | -0.098 | -0.3997 | Yes |
| 215 | FAM43A | FAM43A | 20087 | -0.098 | -0.3980 | Yes |
| 216 | IER3 | IER3 | 20182 | -0.102 | -0.3998 | Yes |
| 217 | NBL1 | NBL1 | 20197 | -0.103 | -0.3982 | Yes |
| 218 | ABR | ABR | 20266 | -0.105 | -0.3989 | Yes |
| 219 | NUDT4 | NUDT4 | 20283 | -0.106 | -0.3973 | Yes |
| 220 | CLDN4 | CLDN4 | 20291 | -0.107 | -0.3954 | Yes |
| 221 | FZD8 | FZD8 | 20308 | -0.107 | -0.3939 | Yes |
| 222 | FAM131B | FAM131B | 20375 | -0.111 | -0.3943 | Yes |
| 223 | SYT12 | SYT12 | 20616 | -0.124 | -0.4017 | Yes |
| 224 | NAB2 | NAB2 | 20634 | -0.124 | -0.3998 | Yes |
| 225 | IGFBP3 | IGFBP3 | 20754 | -0.131 | -0.4020 | Yes |
| 226 | DACT1 | DACT1 | 20763 | -0.132 | -0.3996 | Yes |
| 227 | FZD5 | FZD5 | 20828 | -0.135 | -0.3995 | Yes |
| 228 | SDC4 | SDC4 | 20885 | -0.139 | -0.3989 | Yes |
| 229 | CTSS | CTSS | 20979 | -0.149 | -0.3997 | Yes |
| 230 | LRRN3 | LRRN3 | 21008 | -0.153 | -0.3977 | Yes |
| 231 | CAMK2D | CAMK2D | 21027 | -0.154 | -0.3952 | Yes |
| 232 | IL1RAP | IL1RAP | 21028 | -0.154 | -0.3920 | Yes |
| 233 | DTX3L | DTX3L | 21046 | -0.156 | -0.3895 | Yes |
| 234 | PARP9 | PARP9 | 21047 | -0.156 | -0.3862 | Yes |
| 235 | CAPN5 | CAPN5 | 21078 | -0.159 | -0.3842 | Yes |
| 236 | STOM | STOM | 21124 | -0.163 | -0.3827 | Yes |
| 237 | EMP1 | EMP1 | 21196 | -0.170 | -0.3821 | Yes |
| 238 | GABRA5 | GABRA5 | 21210 | -0.172 | -0.3791 | Yes |
| 239 | L1CAM | L1CAM | 21234 | -0.174 | -0.3764 | Yes |
| 240 | SPSB1 | SPSB1 | 21522 | -0.205 | -0.3841 | Yes |
| 241 | LRP2 | LRP2 | 21548 | -0.208 | -0.3808 | Yes |
| 242 | FGF2 | FGF2 | 21561 | -0.209 | -0.3769 | Yes |
| 243 | SH2B3 | SH2B3 | 21658 | -0.219 | -0.3764 | Yes |
| 244 | CITED4 | CITED4 | 21737 | -0.230 | -0.3748 | Yes |
| 245 | IFIH1 | IFIH1 | 21835 | -0.244 | -0.3738 | Yes |
| 246 | BMP5 | BMP5 | 21844 | -0.245 | -0.3690 | Yes |
| 247 | EMILIN2 | EMILIN2 | 21848 | -0.246 | -0.3641 | Yes |
| 248 | CYP1A1 | CYP1A1 | 21869 | -0.249 | -0.3597 | Yes |
| 249 | A2M | A2M | 21880 | -0.251 | -0.3549 | Yes |
| 250 | CDC42EP5 | CDC42EP5 | 21895 | -0.253 | -0.3503 | Yes |
| 251 | GFRA1 | GFRA1 | 21969 | -0.266 | -0.3478 | Yes |
| 252 | BACE2 | BACE2 | 22035 | -0.275 | -0.3448 | Yes |
| 253 | SLC2A3 | SLC2A3 | 22057 | -0.278 | -0.3399 | Yes |
| 254 | TNFRSF12A | TNFRSF12A | 22148 | -0.293 | -0.3375 | Yes |
| 255 | VGF | VGF | 22175 | -0.298 | -0.3324 | Yes |
| 256 | NR4A3 | NR4A3 | 22302 | -0.321 | -0.3310 | Yes |
| 257 | SYNPO | SYNPO | 22326 | -0.327 | -0.3251 | Yes |
| 258 | TRPM8 | TRPM8 | 22449 | -0.349 | -0.3230 | Yes |
| 259 | MICALL2 | MICALL2 | 23001 | -0.362 | -0.3383 | Yes |
| 260 | LARP6 | LARP6 | 23042 | -0.371 | -0.3323 | Yes |
| 261 | ADAMTS14 | ADAMTS14 | 23098 | -0.380 | -0.3267 | Yes |
| 262 | CDKN2B | CDKN2B | 23135 | -0.392 | -0.3200 | Yes |
| 263 | SPP1 | SPP1 | 23241 | -0.405 | -0.3160 | Yes |
| 264 | ASPHD1 | ASPHD1 | 23321 | -0.424 | -0.3105 | Yes |
| 265 | CSRP2 | CSRP2 | 23365 | -0.436 | -0.3032 | Yes |
| 266 | EGFLAM | EGFLAM | 23396 | -0.445 | -0.2952 | Yes |
| 267 | RGS16 | RGS16 | 23418 | -0.454 | -0.2867 | Yes |
| 268 | IL4I1 | IL4I1 | 23432 | -0.459 | -0.2777 | Yes |
| 269 | PERP | PERP | 23434 | -0.459 | -0.2682 | Yes |
| 270 | ASB9 | ASB9 | 23592 | -0.524 | -0.2638 | Yes |
| 271 | DOC2B | DOC2B | 23605 | -0.530 | -0.2533 | Yes |
| 272 | PGM2L1 | PGM2L1 | 23652 | -0.545 | -0.2439 | Yes |
| 273 | TNFRSF9 | TNFRSF9 | 23679 | -0.558 | -0.2335 | Yes |
| 274 | PTPRE | PTPRE | 23712 | -0.582 | -0.2227 | Yes |
| 275 | PHF24 | PHF24 | 23716 | -0.585 | -0.2107 | Yes |
| 276 | ARHGEF16 | ARHGEF16 | 23807 | -0.638 | -0.2012 | Yes |
| 277 | FADS3 | FADS3 | 23982 | -0.718 | -0.1935 | Yes |
| 278 | AHR | AHR | 24012 | -0.750 | -0.1792 | Yes |
| 279 | ISG20 | ISG20 | 24023 | -0.758 | -0.1639 | Yes |
| 280 | MAOB | MAOB | 24025 | -0.758 | -0.1482 | Yes |
| 281 | CD83 | CD83 | 24121 | -0.915 | -0.1331 | Yes |
| 282 | FAM129A | FAM129A | 24138 | -0.933 | -0.1144 | Yes |
| 283 | ETNK2 | ETNK2 | 24158 | -0.983 | -0.0948 | Yes |
| 284 | SGPP2 | SGPP2 | 24322 | -1.513 | -0.0702 | Yes |
| 285 | NRG1 | NRG1 | 24335 | -1.630 | -0.0369 | Yes |
| 286 | LHX9 | LHX9 | 24365 | -1.883 | 0.0010 | Yes |

