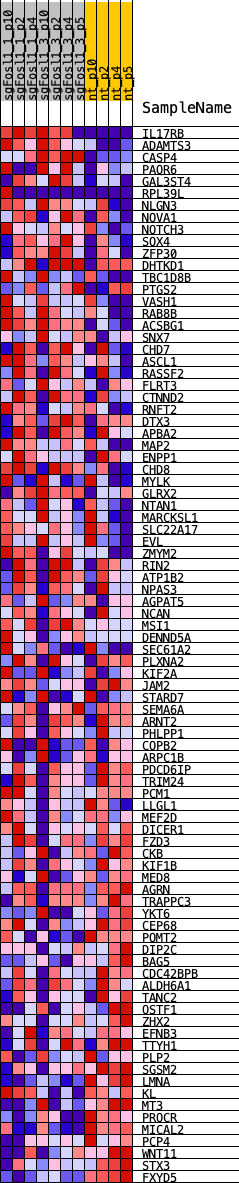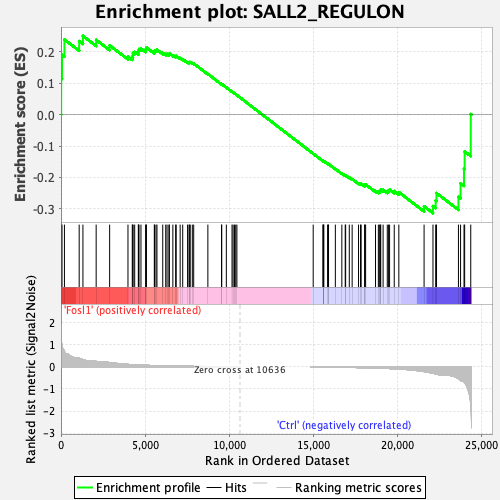
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | SALL2_REGULON |
| Enrichment Score (ES) | -0.31376374 |
| Normalized Enrichment Score (NES) | -1.2332503 |
| Nominal p-value | 0.06042885 |
| FDR q-value | 0.26928315 |
| FWER p-Value | 0.838 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IL17RB | IL17RB | 11 | 1.472 | 0.1154 | No |
| 2 | ADAMTS3 | ADAMTS3 | 59 | 0.982 | 0.1908 | No |
| 3 | CASP4 | CASP4 | 204 | 0.693 | 0.2394 | No |
| 4 | PAQR6 | PAQR6 | 1080 | 0.393 | 0.2343 | No |
| 5 | GAL3ST4 | GAL3ST4 | 1303 | 0.328 | 0.2510 | No |
| 6 | RPL39L | RPL39L | 2089 | 0.249 | 0.2383 | No |
| 7 | NLGN3 | NLGN3 | 2890 | 0.197 | 0.2209 | No |
| 8 | NOVA1 | NOVA1 | 3979 | 0.112 | 0.1849 | No |
| 9 | NOTCH3 | NOTCH3 | 4234 | 0.101 | 0.1824 | No |
| 10 | SOX4 | SOX4 | 4269 | 0.100 | 0.1889 | No |
| 11 | ZFP30 | ZFP30 | 4276 | 0.099 | 0.1964 | No |
| 12 | DHTKD1 | DHTKD1 | 4378 | 0.094 | 0.1997 | No |
| 13 | TBC1D8B | TBC1D8B | 4596 | 0.084 | 0.1974 | No |
| 14 | PTGS2 | PTGS2 | 4602 | 0.084 | 0.2038 | No |
| 15 | VASH1 | VASH1 | 4636 | 0.083 | 0.2090 | No |
| 16 | RAB8B | RAB8B | 4751 | 0.080 | 0.2106 | No |
| 17 | ACSBG1 | ACSBG1 | 5032 | 0.072 | 0.2047 | No |
| 18 | SNX7 | SNX7 | 5046 | 0.071 | 0.2098 | No |
| 19 | CHD7 | CHD7 | 5075 | 0.070 | 0.2142 | No |
| 20 | ASCL1 | ASCL1 | 5543 | 0.059 | 0.1996 | No |
| 21 | RASSF2 | RASSF2 | 5555 | 0.059 | 0.2038 | No |
| 22 | FLRT3 | FLRT3 | 5641 | 0.057 | 0.2048 | No |
| 23 | CTNND2 | CTNND2 | 5701 | 0.056 | 0.2068 | No |
| 24 | RNFT2 | RNFT2 | 6045 | 0.050 | 0.1966 | No |
| 25 | DTX3 | DTX3 | 6222 | 0.047 | 0.1930 | No |
| 26 | APBA2 | APBA2 | 6300 | 0.046 | 0.1935 | No |
| 27 | MAP2 | MAP2 | 6412 | 0.044 | 0.1923 | No |
| 28 | ENPP1 | ENPP1 | 6438 | 0.043 | 0.1947 | No |
| 29 | CHD8 | CHD8 | 6646 | 0.040 | 0.1894 | No |
| 30 | MYLK | MYLK | 6812 | 0.038 | 0.1856 | No |
| 31 | GLRX2 | GLRX2 | 6843 | 0.038 | 0.1873 | No |
| 32 | NTAN1 | NTAN1 | 7079 | 0.035 | 0.1803 | No |
| 33 | MARCKSL1 | MARCKSL1 | 7231 | 0.033 | 0.1767 | No |
| 34 | SLC22A17 | SLC22A17 | 7520 | 0.029 | 0.1672 | No |
| 35 | EVL | EVL | 7611 | 0.028 | 0.1657 | No |
| 36 | ZMYM2 | ZMYM2 | 7623 | 0.028 | 0.1674 | No |
| 37 | RIN2 | RIN2 | 7678 | 0.027 | 0.1673 | No |
| 38 | ATP1B2 | ATP1B2 | 7802 | 0.026 | 0.1643 | No |
| 39 | NPAS3 | NPAS3 | 7884 | 0.025 | 0.1630 | No |
| 40 | AGPAT5 | AGPAT5 | 8726 | 0.017 | 0.1297 | No |
| 41 | NCAN | NCAN | 9540 | 0.010 | 0.0970 | No |
| 42 | MSI1 | MSI1 | 9548 | 0.009 | 0.0974 | No |
| 43 | DENND5A | DENND5A | 9823 | 0.007 | 0.0867 | No |
| 44 | SEC61A2 | SEC61A2 | 10161 | 0.004 | 0.0732 | No |
| 45 | PLXNA2 | PLXNA2 | 10225 | 0.003 | 0.0708 | No |
| 46 | KIF2A | KIF2A | 10307 | 0.003 | 0.0677 | No |
| 47 | JAM2 | JAM2 | 10381 | 0.002 | 0.0649 | No |
| 48 | STARD7 | STARD7 | 10450 | 0.002 | 0.0622 | No |
| 49 | SEMA6A | SEMA6A | 14984 | -0.001 | -0.1242 | No |
| 50 | ARNT2 | ARNT2 | 15571 | -0.006 | -0.1478 | No |
| 51 | PHLPP1 | PHLPP1 | 15580 | -0.006 | -0.1476 | No |
| 52 | COPB2 | COPB2 | 15582 | -0.007 | -0.1472 | No |
| 53 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.1480 | No |
| 54 | PDCD6IP | PDCD6IP | 15840 | -0.009 | -0.1565 | No |
| 55 | TRIM24 | TRIM24 | 15878 | -0.009 | -0.1572 | No |
| 56 | PCM1 | PCM1 | 15895 | -0.009 | -0.1572 | No |
| 57 | LLGL1 | LLGL1 | 16300 | -0.014 | -0.1727 | No |
| 58 | MEF2D | MEF2D | 16686 | -0.017 | -0.1872 | No |
| 59 | DICER1 | DICER1 | 16887 | -0.020 | -0.1939 | No |
| 60 | FZD3 | FZD3 | 16912 | -0.020 | -0.1933 | No |
| 61 | CKB | CKB | 17128 | -0.023 | -0.2003 | No |
| 62 | KIF1B | KIF1B | 17298 | -0.025 | -0.2054 | No |
| 63 | MED8 | MED8 | 17677 | -0.030 | -0.2186 | No |
| 64 | AGRN | AGRN | 17795 | -0.031 | -0.2209 | No |
| 65 | TRAPPC3 | TRAPPC3 | 17832 | -0.032 | -0.2199 | No |
| 66 | YKT6 | YKT6 | 18042 | -0.035 | -0.2257 | No |
| 67 | CEP68 | CEP68 | 18052 | -0.035 | -0.2233 | No |
| 68 | POMT2 | POMT2 | 18099 | -0.036 | -0.2223 | No |
| 69 | DIP2C | DIP2C | 18687 | -0.048 | -0.2428 | No |
| 70 | BAG5 | BAG5 | 18856 | -0.051 | -0.2456 | No |
| 71 | CDC42BPB | CDC42BPB | 18934 | -0.054 | -0.2446 | No |
| 72 | ALDH6A1 | ALDH6A1 | 18972 | -0.055 | -0.2418 | No |
| 73 | TANC2 | TANC2 | 18997 | -0.055 | -0.2385 | No |
| 74 | OSTF1 | OSTF1 | 19137 | -0.059 | -0.2395 | No |
| 75 | ZHX2 | ZHX2 | 19385 | -0.068 | -0.2443 | No |
| 76 | EFNB3 | EFNB3 | 19453 | -0.070 | -0.2416 | No |
| 77 | TTYH1 | TTYH1 | 19522 | -0.072 | -0.2387 | No |
| 78 | PLP2 | PLP2 | 19799 | -0.084 | -0.2434 | No |
| 79 | SGSM2 | SGSM2 | 20072 | -0.097 | -0.2469 | No |
| 80 | LMNA | LMNA | 21572 | -0.210 | -0.2921 | Yes |
| 81 | KL | KL | 22100 | -0.285 | -0.2913 | Yes |
| 82 | MT3 | MT3 | 22262 | -0.313 | -0.2733 | Yes |
| 83 | PROCR | PROCR | 22311 | -0.323 | -0.2499 | Yes |
| 84 | MICAL2 | MICAL2 | 23614 | -0.532 | -0.2615 | Yes |
| 85 | PCP4 | PCP4 | 23740 | -0.609 | -0.2187 | Yes |
| 86 | WNT11 | WNT11 | 23945 | -0.695 | -0.1725 | Yes |
| 87 | STX3 | STX3 | 23983 | -0.720 | -0.1173 | Yes |
| 88 | FXYD5 | FXYD5 | 24345 | -1.702 | 0.0018 | Yes |