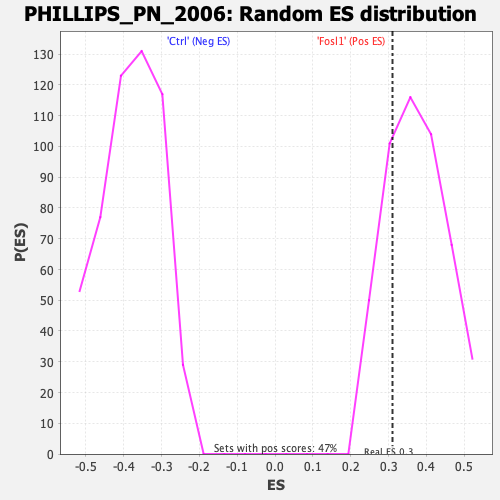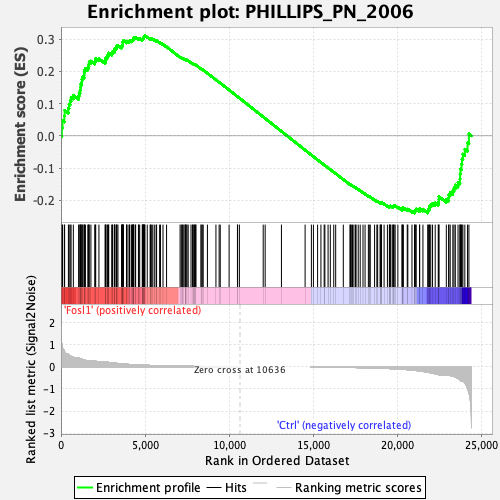
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Fosl1 |
| GeneSet | PHILLIPS_PN_2006 |
| Enrichment Score (ES) | 0.31006643 |
| Normalized Enrichment Score (NES) | 0.8338828 |
| Nominal p-value | 0.7446808 |
| FDR q-value | 0.7091927 |
| FWER p-Value | 0.994 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GABRA3 | GABRA3 | 52 | 1.006 | 0.0252 | Yes |
| 2 | RASGEF1C | RASGEF1C | 90 | 0.869 | 0.0473 | Yes |
| 3 | CNTN3 | CNTN3 | 197 | 0.700 | 0.0619 | Yes |
| 4 | ARPP21 | ARPP21 | 216 | 0.672 | 0.0794 | Yes |
| 5 | FAIM2 | FAIM2 | 439 | 0.562 | 0.0855 | Yes |
| 6 | MMP16 | MMP16 | 479 | 0.536 | 0.0985 | Yes |
| 7 | NKAIN4 | NKAIN4 | 544 | 0.496 | 0.1093 | Yes |
| 8 | OMG | OMG | 598 | 0.473 | 0.1199 | Yes |
| 9 | GRIA4 | GRIA4 | 733 | 0.432 | 0.1261 | Yes |
| 10 | PDK4 | PDK4 | 1050 | 0.401 | 0.1239 | Yes |
| 11 | DUSP26 | DUSP26 | 1078 | 0.393 | 0.1335 | Yes |
| 12 | C1QL1 | C1QL1 | 1134 | 0.374 | 0.1414 | Yes |
| 13 | GABBR2 | GABBR2 | 1158 | 0.366 | 0.1504 | Yes |
| 14 | DPP10 | DPP10 | 1167 | 0.364 | 0.1600 | Yes |
| 15 | SLITRK5 | SLITRK5 | 1234 | 0.346 | 0.1666 | Yes |
| 16 | HEY2 | HEY2 | 1241 | 0.344 | 0.1757 | Yes |
| 17 | NAP1L3 | NAP1L3 | 1289 | 0.331 | 0.1828 | Yes |
| 18 | FGF13 | FGF13 | 1375 | 0.311 | 0.1877 | Yes |
| 19 | FGF14 | FGF14 | 1379 | 0.310 | 0.1960 | Yes |
| 20 | USH1C | USH1C | 1392 | 0.307 | 0.2039 | Yes |
| 21 | SSTR1 | SSTR1 | 1451 | 0.296 | 0.2095 | Yes |
| 22 | SGCG | SGCG | 1594 | 0.274 | 0.2111 | Yes |
| 23 | RASL10A | RASL10A | 1643 | 0.264 | 0.2163 | Yes |
| 24 | TMEM100 | TMEM100 | 1659 | 0.262 | 0.2228 | Yes |
| 25 | DSCAM | DSCAM | 1671 | 0.261 | 0.2294 | Yes |
| 26 | RPRM | RPRM | 1777 | 0.252 | 0.2320 | Yes |
| 27 | RIPPLY2 | RIPPLY2 | 2008 | 0.250 | 0.2292 | Yes |
| 28 | FGF12 | FGF12 | 2037 | 0.249 | 0.2348 | Yes |
| 29 | HS3ST4 | HS3ST4 | 2062 | 0.249 | 0.2406 | Yes |
| 30 | P2RY13 | P2RY13 | 2253 | 0.245 | 0.2394 | Yes |
| 31 | GAD1 | GAD1 | 2618 | 0.233 | 0.2307 | Yes |
| 32 | OPCML | OPCML | 2646 | 0.230 | 0.2358 | Yes |
| 33 | NDRG2 | NDRG2 | 2656 | 0.228 | 0.2417 | Yes |
| 34 | FXYD6 | FXYD6 | 2737 | 0.216 | 0.2442 | Yes |
| 35 | SNRPN | SNRPN | 2755 | 0.213 | 0.2493 | Yes |
| 36 | DLL3 | DLL3 | 2822 | 0.204 | 0.2521 | Yes |
| 37 | DSCAML1 | DSCAML1 | 2837 | 0.202 | 0.2570 | Yes |
| 38 | LUZP2 | LUZP2 | 3016 | 0.183 | 0.2546 | Yes |
| 39 | RGS9 | RGS9 | 3049 | 0.181 | 0.2582 | Yes |
| 40 | LGR5 | LGR5 | 3072 | 0.178 | 0.2622 | Yes |
| 41 | GNAL | GNAL | 3174 | 0.169 | 0.2626 | Yes |
| 42 | KCTD4 | KCTD4 | 3184 | 0.168 | 0.2668 | Yes |
| 43 | GPR158 | GPR158 | 3196 | 0.167 | 0.2708 | Yes |
| 44 | CMTM5 | CMTM5 | 3278 | 0.160 | 0.2719 | Yes |
| 45 | SLC1A1 | SLC1A1 | 3310 | 0.158 | 0.2749 | Yes |
| 46 | NEU4 | NEU4 | 3313 | 0.158 | 0.2791 | Yes |
| 47 | PTGDS | PTGDS | 3383 | 0.151 | 0.2803 | Yes |
| 48 | CECR6 | CECR6 | 3596 | 0.133 | 0.2752 | Yes |
| 49 | FRY | FRY | 3609 | 0.133 | 0.2783 | Yes |
| 50 | PDE2A | PDE2A | 3648 | 0.131 | 0.2803 | Yes |
| 51 | MN1 | MN1 | 3669 | 0.130 | 0.2830 | Yes |
| 52 | TAL1 | TAL1 | 3672 | 0.130 | 0.2864 | Yes |
| 53 | KSR2 | KSR2 | 3673 | 0.130 | 0.2900 | Yes |
| 54 | DOK6 | DOK6 | 3684 | 0.129 | 0.2931 | Yes |
| 55 | CBX7 | CBX7 | 3708 | 0.128 | 0.2956 | Yes |
| 56 | EPHB1 | EPHB1 | 3890 | 0.116 | 0.2912 | Yes |
| 57 | IL17D | IL17D | 3905 | 0.115 | 0.2938 | Yes |
| 58 | DLL1 | DLL1 | 3982 | 0.112 | 0.2937 | Yes |
| 59 | SHD | SHD | 4071 | 0.108 | 0.2930 | Yes |
| 60 | PLEKHB1 | PLEKHB1 | 4084 | 0.107 | 0.2954 | Yes |
| 61 | EHD3 | EHD3 | 4183 | 0.103 | 0.2942 | Yes |
| 62 | CSMD3 | CSMD3 | 4223 | 0.102 | 0.2953 | Yes |
| 63 | SMOC1 | SMOC1 | 4249 | 0.100 | 0.2970 | Yes |
| 64 | SOX8 | SOX8 | 4294 | 0.099 | 0.2979 | Yes |
| 65 | RUNDC3A | RUNDC3A | 4296 | 0.099 | 0.3005 | Yes |
| 66 | CNTN1 | CNTN1 | 4306 | 0.098 | 0.3028 | Yes |
| 67 | FUT9 | FUT9 | 4325 | 0.097 | 0.3047 | Yes |
| 68 | GABRB3 | GABRB3 | 4414 | 0.093 | 0.3035 | Yes |
| 69 | OLIG1 | OLIG1 | 4434 | 0.092 | 0.3052 | Yes |
| 70 | SORBS1 | SORBS1 | 4601 | 0.084 | 0.3007 | Yes |
| 71 | SOX6 | SOX6 | 4622 | 0.083 | 0.3021 | Yes |
| 72 | GRID1 | GRID1 | 4676 | 0.082 | 0.3021 | Yes |
| 73 | ATP6V1G2 | ATP6V1G2 | 4828 | 0.077 | 0.2980 | Yes |
| 74 | ABLIM1 | ABLIM1 | 4848 | 0.077 | 0.2993 | Yes |
| 75 | DNAJC12 | DNAJC12 | 4852 | 0.076 | 0.3012 | Yes |
| 76 | B3GAT1 | B3GAT1 | 4871 | 0.076 | 0.3026 | Yes |
| 77 | JPH4 | JPH4 | 4904 | 0.075 | 0.3033 | Yes |
| 78 | RIMS2 | RIMS2 | 4913 | 0.075 | 0.3050 | Yes |
| 79 | PDK2 | PDK2 | 4927 | 0.075 | 0.3065 | Yes |
| 80 | MAPT | MAPT | 4940 | 0.074 | 0.3080 | Yes |
| 81 | NTRK2 | NTRK2 | 4961 | 0.074 | 0.3092 | Yes |
| 82 | CALCRL | CALCRL | 4988 | 0.073 | 0.3101 | Yes |
| 83 | OLIG2 | OLIG2 | 5125 | 0.069 | 0.3063 | No |
| 84 | DNM3 | DNM3 | 5295 | 0.065 | 0.3011 | No |
| 85 | GAB2 | GAB2 | 5321 | 0.064 | 0.3018 | No |
| 86 | ADCY2 | ADCY2 | 5367 | 0.063 | 0.3016 | No |
| 87 | TMOD2 | TMOD2 | 5450 | 0.061 | 0.2999 | No |
| 88 | ASCL1 | ASCL1 | 5543 | 0.059 | 0.2977 | No |
| 89 | GRIK4 | GRIK4 | 5650 | 0.057 | 0.2949 | No |
| 90 | PSD | PSD | 5680 | 0.056 | 0.2952 | No |
| 91 | APOE | APOE | 5860 | 0.053 | 0.2892 | No |
| 92 | SEZ6L | SEZ6L | 5910 | 0.052 | 0.2886 | No |
| 93 | RPL37 | RPL37 | 6060 | 0.050 | 0.2838 | No |
| 94 | PKNOX2 | PKNOX2 | 6272 | 0.046 | 0.2763 | No |
| 95 | IKZF5 | IKZF5 | 7076 | 0.035 | 0.2441 | No |
| 96 | GLUD1 | GLUD1 | 7146 | 0.034 | 0.2421 | No |
| 97 | SLITRK2 | SLITRK2 | 7196 | 0.033 | 0.2410 | No |
| 98 | RTN1 | RTN1 | 7229 | 0.033 | 0.2406 | No |
| 99 | ALDOC | ALDOC | 7295 | 0.032 | 0.2387 | No |
| 100 | FBXL15 | FBXL15 | 7377 | 0.031 | 0.2362 | No |
| 101 | RPL13 | RPL13 | 7379 | 0.031 | 0.2370 | No |
| 102 | KCNB1 | KCNB1 | 7420 | 0.030 | 0.2362 | No |
| 103 | FAM19A5 | FAM19A5 | 7428 | 0.030 | 0.2367 | No |
| 104 | ENHO | ENHO | 7486 | 0.030 | 0.2352 | No |
| 105 | ZCCHC24 | ZCCHC24 | 7556 | 0.029 | 0.2331 | No |
| 106 | PHYHIPL | PHYHIPL | 7724 | 0.027 | 0.2269 | No |
| 107 | GGTA1 | GGTA1 | 7809 | 0.026 | 0.2242 | No |
| 108 | ABHD6 | ABHD6 | 7846 | 0.026 | 0.2234 | No |
| 109 | PCSK1N | PCSK1N | 7850 | 0.026 | 0.2239 | No |
| 110 | BCAN | BCAN | 7937 | 0.025 | 0.2211 | No |
| 111 | ASB13 | ASB13 | 7945 | 0.025 | 0.2214 | No |
| 112 | FAM13C | FAM13C | 7997 | 0.024 | 0.2200 | No |
| 113 | ALDH5A1 | ALDH5A1 | 8018 | 0.024 | 0.2198 | No |
| 114 | THRA | THRA | 8319 | 0.020 | 0.2079 | No |
| 115 | GPRC5B | GPRC5B | 8329 | 0.020 | 0.2081 | No |
| 116 | NRSN1 | NRSN1 | 8423 | 0.019 | 0.2048 | No |
| 117 | TIMP4 | TIMP4 | 8442 | 0.019 | 0.2046 | No |
| 118 | GABBR1 | GABBR1 | 8699 | 0.017 | 0.1944 | No |
| 119 | LRRC4 | LRRC4 | 9205 | 0.012 | 0.1739 | No |
| 120 | NUMA1 | NUMA1 | 9395 | 0.011 | 0.1664 | No |
| 121 | NCAM1 | NCAM1 | 9461 | 0.010 | 0.1640 | No |
| 122 | TPCN2 | TPCN2 | 9988 | 0.005 | 0.1424 | No |
| 123 | ZC3H12B | ZC3H12B | 10484 | 0.001 | 0.1219 | No |
| 124 | MPPE1 | MPPE1 | 10491 | 0.001 | 0.1217 | No |
| 125 | ANKS1B | ANKS1B | 10598 | 0.000 | 0.1173 | No |
| 126 | AJAP1 | AJAP1 | 12013 | 0.000 | 0.0588 | No |
| 127 | CDR1 | CDR1 | 12130 | 0.000 | 0.0540 | No |
| 128 | KLRC3 | KLRC3 | 13097 | 0.000 | 0.0141 | No |
| 129 | TMLHE | TMLHE | 14503 | 0.000 | -0.0440 | No |
| 130 | PID1 | PID1 | 14876 | -0.000 | -0.0594 | No |
| 131 | CRTC1 | CRTC1 | 14997 | -0.002 | -0.0644 | No |
| 132 | HDAC5 | HDAC5 | 15246 | -0.004 | -0.0745 | No |
| 133 | MAPK8IP2 | MAPK8IP2 | 15443 | -0.005 | -0.0825 | No |
| 134 | BMP2 | BMP2 | 15635 | -0.007 | -0.0902 | No |
| 135 | RAC3 | RAC3 | 15678 | -0.008 | -0.0917 | No |
| 136 | CSDC2 | CSDC2 | 15868 | -0.009 | -0.0993 | No |
| 137 | SH3GL2 | SH3GL2 | 16001 | -0.011 | -0.1044 | No |
| 138 | AP2B1 | AP2B1 | 16205 | -0.013 | -0.1125 | No |
| 139 | PCSK6 | PCSK6 | 16320 | -0.014 | -0.1168 | No |
| 140 | MARCH8 | MARCH8 | 16773 | -0.018 | -0.1350 | No |
| 141 | PARD3 | PARD3 | 17167 | -0.023 | -0.1507 | No |
| 142 | ARL3 | ARL3 | 17200 | -0.024 | -0.1513 | No |
| 143 | TNKS2 | TNKS2 | 17249 | -0.024 | -0.1527 | No |
| 144 | FAM155A | FAM155A | 17319 | -0.025 | -0.1549 | No |
| 145 | KIF5A | KIF5A | 17402 | -0.026 | -0.1575 | No |
| 146 | CCNK | CCNK | 17493 | -0.027 | -0.1605 | No |
| 147 | HLF | HLF | 17531 | -0.027 | -0.1613 | No |
| 148 | PRKCZ | PRKCZ | 17659 | -0.029 | -0.1658 | No |
| 149 | SNAP91 | SNAP91 | 17777 | -0.031 | -0.1698 | No |
| 150 | KIF1A | KIF1A | 17943 | -0.034 | -0.1757 | No |
| 151 | GALNT13 | GALNT13 | 18066 | -0.036 | -0.1798 | No |
| 152 | KIF21B | KIF21B | 18262 | -0.039 | -0.1868 | No |
| 153 | PKP4 | PKP4 | 18315 | -0.040 | -0.1878 | No |
| 154 | SLC1A4 | SLC1A4 | 18374 | -0.041 | -0.1891 | No |
| 155 | P2RX7 | P2RX7 | 18631 | -0.046 | -0.1984 | No |
| 156 | FERMT1 | FERMT1 | 18767 | -0.050 | -0.2027 | No |
| 157 | HIP1R | HIP1R | 18813 | -0.050 | -0.2032 | No |
| 158 | SGSM1 | SGSM1 | 18968 | -0.054 | -0.2081 | No |
| 159 | OVOL1 | OVOL1 | 18969 | -0.054 | -0.2066 | No |
| 160 | GNAO1 | GNAO1 | 19021 | -0.056 | -0.2072 | No |
| 161 | NET1 | NET1 | 19044 | -0.056 | -0.2066 | No |
| 162 | PLCB1 | PLCB1 | 19196 | -0.061 | -0.2111 | No |
| 163 | ATRNL1 | ATRNL1 | 19394 | -0.068 | -0.2174 | No |
| 164 | ID4 | ID4 | 19509 | -0.072 | -0.2202 | No |
| 165 | TTYH1 | TTYH1 | 19522 | -0.072 | -0.2187 | No |
| 166 | FSD1 | FSD1 | 19550 | -0.073 | -0.2178 | No |
| 167 | ELMO1 | ELMO1 | 19587 | -0.075 | -0.2173 | No |
| 168 | REPS2 | REPS2 | 19681 | -0.079 | -0.2190 | No |
| 169 | DTX4 | DTX4 | 19734 | -0.081 | -0.2190 | No |
| 170 | GRIA2 | GRIA2 | 19753 | -0.082 | -0.2175 | No |
| 171 | BEND7 | BEND7 | 19791 | -0.084 | -0.2167 | No |
| 172 | NTN4 | NTN4 | 19853 | -0.087 | -0.2169 | No |
| 173 | KCNQ5 | KCNQ5 | 20014 | -0.094 | -0.2210 | No |
| 174 | SEC31B | SEC31B | 20270 | -0.105 | -0.2286 | No |
| 175 | STOX1 | STOX1 | 20272 | -0.105 | -0.2258 | No |
| 176 | WDR86 | WDR86 | 20278 | -0.106 | -0.2232 | No |
| 177 | SUSD5 | SUSD5 | 20342 | -0.109 | -0.2228 | No |
| 178 | SCG3 | SCG3 | 20572 | -0.121 | -0.2290 | No |
| 179 | SCN3A | SCN3A | 20606 | -0.123 | -0.2270 | No |
| 180 | SLC25A21 | SLC25A21 | 20853 | -0.138 | -0.2335 | No |
| 181 | MAF | MAF | 21003 | -0.152 | -0.2355 | No |
| 182 | TPM1 | TPM1 | 21017 | -0.153 | -0.2319 | No |
| 183 | KCNN3 | KCNN3 | 21070 | -0.158 | -0.2297 | No |
| 184 | SMAD9 | SMAD9 | 21102 | -0.161 | -0.2266 | No |
| 185 | MCF2 | MCF2 | 21305 | -0.183 | -0.2300 | No |
| 186 | TMEM59L | TMEM59L | 21308 | -0.183 | -0.2251 | No |
| 187 | SORCS3 | SORCS3 | 21504 | -0.204 | -0.2276 | No |
| 188 | SLIT1 | SLIT1 | 21788 | -0.236 | -0.2329 | No |
| 189 | FBXO2 | FBXO2 | 21827 | -0.243 | -0.2279 | No |
| 190 | CRYAB | CRYAB | 21884 | -0.252 | -0.2234 | No |
| 191 | GRIA1 | GRIA1 | 21907 | -0.255 | -0.2173 | No |
| 192 | GFRA1 | GFRA1 | 21969 | -0.266 | -0.2126 | No |
| 193 | CALN1 | CALN1 | 22075 | -0.282 | -0.2093 | No |
| 194 | WNT7B | WNT7B | 22233 | -0.308 | -0.2074 | No |
| 195 | FAM110B | FAM110B | 22413 | -0.344 | -0.2055 | No |
| 196 | JPH3 | JPH3 | 22446 | -0.349 | -0.1973 | No |
| 197 | RAB11FIP4 | RAB11FIP4 | 22453 | -0.351 | -0.1881 | No |
| 198 | TRIM31 | TRIM31 | 22893 | -0.356 | -0.1965 | No |
| 199 | RASSF4 | RASSF4 | 23033 | -0.369 | -0.1923 | No |
| 200 | FSTL5 | FSTL5 | 23043 | -0.372 | -0.1825 | No |
| 201 | CYFIP2 | CYFIP2 | 23124 | -0.389 | -0.1753 | No |
| 202 | HSPA12A | HSPA12A | 23281 | -0.415 | -0.1704 | No |
| 203 | SPHKAP | SPHKAP | 23348 | -0.433 | -0.1614 | No |
| 204 | NALCN | NALCN | 23427 | -0.458 | -0.1522 | No |
| 205 | F2 | F2 | 23589 | -0.523 | -0.1446 | No |
| 206 | ABLIM3 | ABLIM3 | 23703 | -0.575 | -0.1337 | No |
| 207 | SSTR2 | SSTR2 | 23713 | -0.582 | -0.1183 | No |
| 208 | FLRT1 | FLRT1 | 23731 | -0.597 | -0.1027 | No |
| 209 | NRG3 | NRG3 | 23798 | -0.637 | -0.0882 | No |
| 210 | ZDHHC22 | ZDHHC22 | 23824 | -0.638 | -0.0719 | No |
| 211 | ACSM5 | ACSM5 | 23878 | -0.650 | -0.0564 | No |
| 212 | NOG | NOG | 23993 | -0.729 | -0.0413 | No |
| 213 | PHACTR3 | PHACTR3 | 24154 | -0.972 | -0.0215 | No |
| 214 | ARHGAP22 | ARHGAP22 | 24236 | -1.146 | 0.0063 | No |