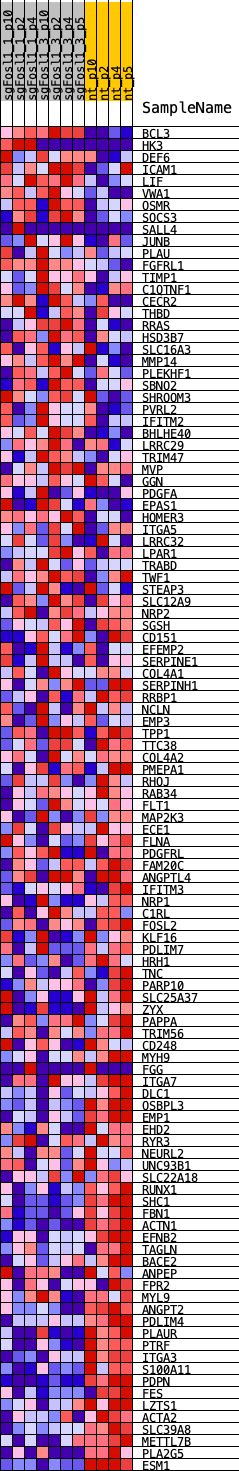
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
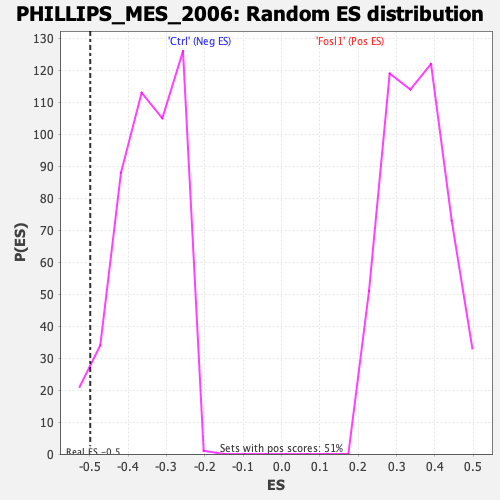
| GeneSet | PHILLIPS_MES_2006 |
| Enrichment Score (ES) | -0.49859944 |
| Normalized Enrichment Score (NES) | -1.435192 |
| Nominal p-value | 0.043032788 |
| FDR q-value | 0.08261187 |
| FWER p-Value | 0.332 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | BCL3 | BCL3 | 167 | 0.756 | 0.0242 | No |
| 2 | HK3 | HK3 | 254 | 0.617 | 0.0459 | No |
| 3 | DEF6 | DEF6 | 278 | 0.607 | 0.0699 | No |
| 4 | ICAM1 | ICAM1 | 511 | 0.517 | 0.0816 | No |
| 5 | LIF | LIF | 668 | 0.445 | 0.0934 | No |
| 6 | VWA1 | VWA1 | 1299 | 0.329 | 0.0809 | No |
| 7 | OSMR | OSMR | 1318 | 0.324 | 0.0935 | No |
| 8 | SOCS3 | SOCS3 | 1528 | 0.284 | 0.0965 | No |
| 9 | SALL4 | SALL4 | 2009 | 0.250 | 0.0870 | No |
| 10 | JUNB | JUNB | 2433 | 0.243 | 0.0795 | No |
| 11 | PLAU | PLAU | 2866 | 0.199 | 0.0699 | No |
| 12 | FGFRL1 | FGFRL1 | 3382 | 0.151 | 0.0549 | No |
| 13 | TIMP1 | TIMP1 | 3528 | 0.139 | 0.0547 | No |
| 14 | C1QTNF1 | C1QTNF1 | 3582 | 0.134 | 0.0580 | No |
| 15 | CECR2 | CECR2 | 3613 | 0.132 | 0.0622 | No |
| 16 | THBD | THBD | 3821 | 0.119 | 0.0586 | No |
| 17 | RRAS | RRAS | 4145 | 0.105 | 0.0496 | No |
| 18 | HSD3B7 | HSD3B7 | 4159 | 0.104 | 0.0533 | No |
| 19 | SLC16A3 | SLC16A3 | 4187 | 0.103 | 0.0564 | No |
| 20 | MMP14 | MMP14 | 4214 | 0.102 | 0.0595 | No |
| 21 | PLEKHF1 | PLEKHF1 | 4218 | 0.102 | 0.0636 | No |
| 22 | SBNO2 | SBNO2 | 4240 | 0.101 | 0.0668 | No |
| 23 | SHROOM3 | SHROOM3 | 4244 | 0.100 | 0.0708 | No |
| 24 | PVRL2 | PVRL2 | 4406 | 0.093 | 0.0680 | No |
| 25 | IFITM2 | IFITM2 | 4895 | 0.075 | 0.0510 | No |
| 26 | BHLHE40 | BHLHE40 | 4962 | 0.074 | 0.0513 | No |
| 27 | LRRC29 | LRRC29 | 4972 | 0.073 | 0.0540 | No |
| 28 | TRIM47 | TRIM47 | 5077 | 0.070 | 0.0526 | No |
| 29 | MVP | MVP | 5446 | 0.061 | 0.0399 | No |
| 30 | GGN | GGN | 5759 | 0.055 | 0.0293 | No |
| 31 | PDGFA | PDGFA | 6104 | 0.049 | 0.0172 | No |
| 32 | EPAS1 | EPAS1 | 6142 | 0.048 | 0.0176 | No |
| 33 | HOMER3 | HOMER3 | 6236 | 0.047 | 0.0157 | No |
| 34 | ITGA5 | ITGA5 | 6940 | 0.036 | -0.0118 | No |
| 35 | LRRC32 | LRRC32 | 7635 | 0.028 | -0.0392 | No |
| 36 | LPAR1 | LPAR1 | 7985 | 0.024 | -0.0526 | No |
| 37 | TRABD | TRABD | 8748 | 0.017 | -0.0833 | No |
| 38 | TWF1 | TWF1 | 8906 | 0.015 | -0.0892 | No |
| 39 | STEAP3 | STEAP3 | 9084 | 0.013 | -0.0959 | No |
| 40 | SLC12A9 | SLC12A9 | 9937 | 0.006 | -0.1308 | No |
| 41 | NRP2 | NRP2 | 9966 | 0.006 | -0.1317 | No |
| 42 | SGSH | SGSH | 10223 | 0.003 | -0.1421 | No |
| 43 | CD151 | CD151 | 10556 | 0.001 | -0.1557 | No |
| 44 | EFEMP2 | EFEMP2 | 15731 | -0.008 | -0.3685 | No |
| 45 | SERPINE1 | SERPINE1 | 15830 | -0.009 | -0.3722 | No |
| 46 | COL4A1 | COL4A1 | 16204 | -0.013 | -0.3870 | No |
| 47 | SERPINH1 | SERPINH1 | 16443 | -0.015 | -0.3962 | No |
| 48 | RRBP1 | RRBP1 | 16591 | -0.016 | -0.4016 | No |
| 49 | NCLN | NCLN | 16751 | -0.018 | -0.4074 | No |
| 50 | EMP3 | EMP3 | 16762 | -0.018 | -0.4071 | No |
| 51 | TPP1 | TPP1 | 16988 | -0.021 | -0.4155 | No |
| 52 | TTC38 | TTC38 | 17107 | -0.022 | -0.4194 | No |
| 53 | COL4A2 | COL4A2 | 17524 | -0.027 | -0.4354 | No |
| 54 | PMEPA1 | PMEPA1 | 17673 | -0.030 | -0.4403 | No |
| 55 | RHOJ | RHOJ | 17845 | -0.032 | -0.4460 | No |
| 56 | RAB34 | RAB34 | 17979 | -0.034 | -0.4501 | No |
| 57 | FLT1 | FLT1 | 18210 | -0.038 | -0.4580 | No |
| 58 | MAP2K3 | MAP2K3 | 18641 | -0.047 | -0.4738 | No |
| 59 | ECE1 | ECE1 | 18718 | -0.048 | -0.4750 | No |
| 60 | FLNA | FLNA | 18719 | -0.048 | -0.4730 | No |
| 61 | PDGFRL | PDGFRL | 18743 | -0.049 | -0.4719 | No |
| 62 | FAM20C | FAM20C | 18775 | -0.050 | -0.4712 | No |
| 63 | ANGPTL4 | ANGPTL4 | 18785 | -0.050 | -0.4695 | No |
| 64 | IFITM3 | IFITM3 | 18918 | -0.053 | -0.4727 | No |
| 65 | NRP1 | NRP1 | 19122 | -0.059 | -0.4787 | No |
| 66 | C1RL | C1RL | 19124 | -0.059 | -0.4763 | No |
| 67 | FOSL2 | FOSL2 | 19180 | -0.061 | -0.4761 | No |
| 68 | KLF16 | KLF16 | 19614 | -0.076 | -0.4908 | No |
| 69 | PDLIM7 | PDLIM7 | 19713 | -0.080 | -0.4916 | No |
| 70 | HRH1 | HRH1 | 19741 | -0.081 | -0.4893 | No |
| 71 | TNC | TNC | 19967 | -0.092 | -0.4948 | Yes |
| 72 | PARP10 | PARP10 | 19983 | -0.093 | -0.4916 | Yes |
| 73 | SLC25A37 | SLC25A37 | 20059 | -0.096 | -0.4908 | Yes |
| 74 | ZYX | ZYX | 20124 | -0.099 | -0.4893 | Yes |
| 75 | PAPPA | PAPPA | 20167 | -0.102 | -0.4869 | Yes |
| 76 | TRIM56 | TRIM56 | 20275 | -0.106 | -0.4870 | Yes |
| 77 | CD248 | CD248 | 20440 | -0.114 | -0.4890 | Yes |
| 78 | MYH9 | MYH9 | 20636 | -0.124 | -0.4920 | Yes |
| 79 | FGG | FGG | 20738 | -0.129 | -0.4908 | Yes |
| 80 | ITGA7 | ITGA7 | 20879 | -0.139 | -0.4909 | Yes |
| 81 | DLC1 | DLC1 | 20939 | -0.145 | -0.4874 | Yes |
| 82 | OSBPL3 | OSBPL3 | 21074 | -0.159 | -0.4864 | Yes |
| 83 | EMP1 | EMP1 | 21196 | -0.170 | -0.4844 | Yes |
| 84 | EHD2 | EHD2 | 21204 | -0.171 | -0.4777 | Yes |
| 85 | RYR3 | RYR3 | 21249 | -0.175 | -0.4723 | Yes |
| 86 | NEURL2 | NEURL2 | 21337 | -0.186 | -0.4682 | Yes |
| 87 | UNC93B1 | UNC93B1 | 21390 | -0.192 | -0.4625 | Yes |
| 88 | SLC22A18 | SLC22A18 | 21596 | -0.212 | -0.4622 | Yes |
| 89 | RUNX1 | RUNX1 | 21632 | -0.216 | -0.4548 | Yes |
| 90 | SHC1 | SHC1 | 21741 | -0.230 | -0.4498 | Yes |
| 91 | FBN1 | FBN1 | 21875 | -0.250 | -0.4450 | Yes |
| 92 | ACTN1 | ACTN1 | 21926 | -0.257 | -0.4365 | Yes |
| 93 | EFNB2 | EFNB2 | 21938 | -0.259 | -0.4264 | Yes |
| 94 | TAGLN | TAGLN | 21965 | -0.265 | -0.4165 | Yes |
| 95 | BACE2 | BACE2 | 22035 | -0.275 | -0.4081 | Yes |
| 96 | ANPEP | ANPEP | 22140 | -0.292 | -0.4004 | Yes |
| 97 | FPR2 | FPR2 | 22222 | -0.305 | -0.3912 | Yes |
| 98 | MYL9 | MYL9 | 22253 | -0.311 | -0.3797 | Yes |
| 99 | ANGPT2 | ANGPT2 | 22343 | -0.330 | -0.3698 | Yes |
| 100 | PDLIM4 | PDLIM4 | 22354 | -0.332 | -0.3566 | Yes |
| 101 | PLAUR | PLAUR | 23251 | -0.408 | -0.3767 | Yes |
| 102 | PTRF | PTRF | 23482 | -0.476 | -0.3667 | Yes |
| 103 | ITGA3 | ITGA3 | 23486 | -0.477 | -0.3472 | Yes |
| 104 | S100A11 | S100A11 | 23510 | -0.487 | -0.3282 | Yes |
| 105 | PDPN | PDPN | 23644 | -0.542 | -0.3114 | Yes |
| 106 | FES | FES | 23965 | -0.703 | -0.2957 | Yes |
| 107 | LZTS1 | LZTS1 | 24090 | -0.849 | -0.2660 | Yes |
| 108 | ACTA2 | ACTA2 | 24120 | -0.914 | -0.2297 | Yes |
| 109 | SLC39A8 | SLC39A8 | 24175 | -1.021 | -0.1900 | Yes |
| 110 | METTL7B | METTL7B | 24238 | -1.149 | -0.1454 | Yes |
| 111 | PLA2G5 | PLA2G5 | 24243 | -1.172 | -0.0974 | Yes |
| 112 | ESM1 | ESM1 | 24386 | -2.518 | 0.0001 | Yes |