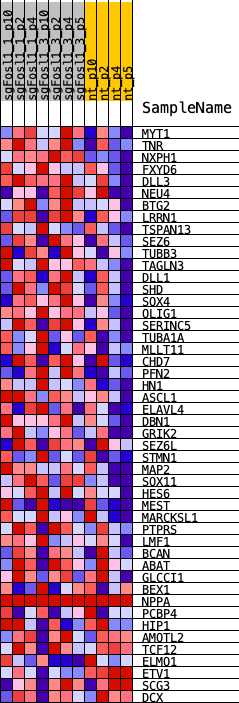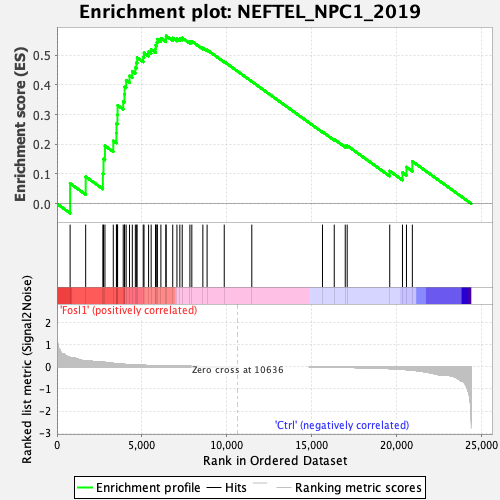
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Fosl1 |
| GeneSet | NEFTEL_NPC1_2019 |
| Enrichment Score (ES) | 0.56468004 |
| Normalized Enrichment Score (NES) | 1.4003693 |
| Nominal p-value | 0.16666667 |
| FDR q-value | 0.40732482 |
| FWER p-Value | 0.375 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MYT1 | MYT1 | 772 | 0.427 | 0.0683 | Yes |
| 2 | TNR | TNR | 1688 | 0.258 | 0.0911 | Yes |
| 3 | NXPH1 | NXPH1 | 2700 | 0.221 | 0.1015 | Yes |
| 4 | FXYD6 | FXYD6 | 2737 | 0.216 | 0.1505 | Yes |
| 5 | DLL3 | DLL3 | 2822 | 0.204 | 0.1948 | Yes |
| 6 | NEU4 | NEU4 | 3313 | 0.158 | 0.2118 | Yes |
| 7 | BTG2 | BTG2 | 3497 | 0.141 | 0.2372 | Yes |
| 8 | LRRN1 | LRRN1 | 3503 | 0.140 | 0.2699 | Yes |
| 9 | TSPAN13 | TSPAN13 | 3558 | 0.136 | 0.2995 | Yes |
| 10 | SEZ6 | SEZ6 | 3567 | 0.135 | 0.3310 | Yes |
| 11 | TUBB3 | TUBB3 | 3898 | 0.116 | 0.3445 | Yes |
| 12 | TAGLN3 | TAGLN3 | 3972 | 0.112 | 0.3678 | Yes |
| 13 | DLL1 | DLL1 | 3982 | 0.112 | 0.3936 | Yes |
| 14 | SHD | SHD | 4071 | 0.108 | 0.4153 | Yes |
| 15 | SOX4 | SOX4 | 4269 | 0.100 | 0.4306 | Yes |
| 16 | OLIG1 | OLIG1 | 4434 | 0.092 | 0.4453 | Yes |
| 17 | SERINC5 | SERINC5 | 4611 | 0.084 | 0.4577 | Yes |
| 18 | TUBA1A | TUBA1A | 4671 | 0.082 | 0.4745 | Yes |
| 19 | MLLT11 | MLLT11 | 4717 | 0.081 | 0.4915 | Yes |
| 20 | CHD7 | CHD7 | 5075 | 0.070 | 0.4933 | Yes |
| 21 | PFN2 | PFN2 | 5122 | 0.069 | 0.5076 | Yes |
| 22 | HN1 | HN1 | 5391 | 0.062 | 0.5112 | Yes |
| 23 | ASCL1 | ASCL1 | 5543 | 0.059 | 0.5188 | Yes |
| 24 | ELAVL4 | ELAVL4 | 5801 | 0.054 | 0.5209 | Yes |
| 25 | DBN1 | DBN1 | 5806 | 0.054 | 0.5334 | Yes |
| 26 | GRIK2 | GRIK2 | 5874 | 0.053 | 0.5430 | Yes |
| 27 | SEZ6L | SEZ6L | 5910 | 0.052 | 0.5537 | Yes |
| 28 | STMN1 | STMN1 | 6116 | 0.049 | 0.5567 | Yes |
| 29 | MAP2 | MAP2 | 6412 | 0.044 | 0.5548 | Yes |
| 30 | SOX11 | SOX11 | 6421 | 0.044 | 0.5647 | Yes |
| 31 | HES6 | HES6 | 6813 | 0.038 | 0.5575 | No |
| 32 | MEST | MEST | 7061 | 0.035 | 0.5555 | No |
| 33 | MARCKSL1 | MARCKSL1 | 7231 | 0.033 | 0.5563 | No |
| 34 | PTPRS | PTPRS | 7371 | 0.031 | 0.5578 | No |
| 35 | LMF1 | LMF1 | 7824 | 0.026 | 0.5453 | No |
| 36 | BCAN | BCAN | 7937 | 0.025 | 0.5465 | No |
| 37 | ABAT | ABAT | 8586 | 0.018 | 0.5241 | No |
| 38 | GLCCI1 | GLCCI1 | 8836 | 0.016 | 0.5175 | No |
| 39 | BEX1 | BEX1 | 9845 | 0.007 | 0.4777 | No |
| 40 | NPPA | NPPA | 11467 | 0.000 | 0.4111 | No |
| 41 | PCBP4 | PCBP4 | 15628 | -0.007 | 0.2418 | No |
| 42 | HIP1 | HIP1 | 16321 | -0.014 | 0.2166 | No |
| 43 | AMOTL2 | AMOTL2 | 16968 | -0.021 | 0.1949 | No |
| 44 | TCF12 | TCF12 | 17083 | -0.022 | 0.1954 | No |
| 45 | ELMO1 | ELMO1 | 19587 | -0.075 | 0.1101 | No |
| 46 | ETV1 | ETV1 | 20337 | -0.109 | 0.1048 | No |
| 47 | SCG3 | SCG3 | 20572 | -0.121 | 0.1234 | No |
| 48 | DCX | DCX | 20918 | -0.142 | 0.1426 | No |