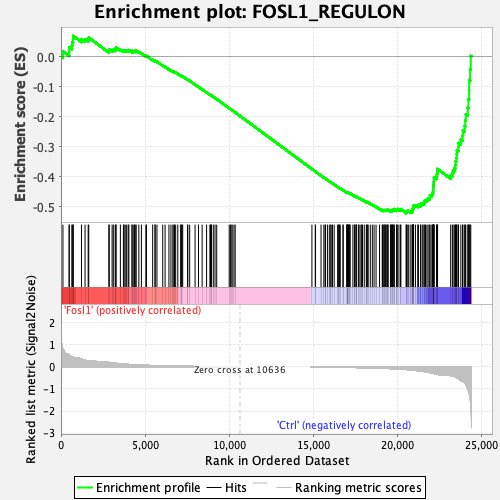
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | FOSL1_REGULON |
| Enrichment Score (ES) | -0.5214364 |
| Normalized Enrichment Score (NES) | -1.5120555 |
| Nominal p-value | 0.034042552 |
| FDR q-value | 0.10422026 |
| FWER p-Value | 0.176 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MOCOS | MOCOS | 115 | 0.810 | 0.0172 | No |
| 2 | VDR | VDR | 484 | 0.533 | 0.0164 | No |
| 3 | KCNN4 | KCNN4 | 487 | 0.528 | 0.0306 | No |
| 4 | IL13RA1 | IL13RA1 | 649 | 0.452 | 0.0362 | No |
| 5 | LIF | LIF | 668 | 0.445 | 0.0475 | No |
| 6 | DRAM1 | DRAM1 | 718 | 0.434 | 0.0572 | No |
| 7 | SRPX2 | SRPX2 | 724 | 0.434 | 0.0687 | No |
| 8 | CYBRD1 | CYBRD1 | 1216 | 0.351 | 0.0579 | No |
| 9 | SH2D4A | SH2D4A | 1430 | 0.300 | 0.0572 | No |
| 10 | MAST1 | MAST1 | 1615 | 0.269 | 0.0569 | No |
| 11 | PTHLH | PTHLH | 1649 | 0.263 | 0.0627 | No |
| 12 | WIPI1 | WIPI1 | 2844 | 0.202 | 0.0187 | No |
| 13 | PLAU | PLAU | 2866 | 0.199 | 0.0232 | No |
| 14 | KCND2 | KCND2 | 3036 | 0.181 | 0.0212 | No |
| 15 | GFPT2 | GFPT2 | 3109 | 0.175 | 0.0229 | No |
| 16 | BFSP1 | BFSP1 | 3221 | 0.165 | 0.0228 | No |
| 17 | CXXC4 | CXXC4 | 3258 | 0.162 | 0.0257 | No |
| 18 | RELB | RELB | 3268 | 0.161 | 0.0297 | No |
| 19 | TIMP1 | TIMP1 | 3528 | 0.139 | 0.0227 | No |
| 20 | CBX7 | CBX7 | 3708 | 0.128 | 0.0188 | No |
| 21 | TNFRSF10B | TNFRSF10B | 3748 | 0.124 | 0.0205 | No |
| 22 | TFPI2 | TFPI2 | 3815 | 0.120 | 0.0210 | No |
| 23 | ADORA2B | ADORA2B | 3899 | 0.116 | 0.0207 | No |
| 24 | DLX2 | DLX2 | 3991 | 0.112 | 0.0200 | No |
| 25 | CDC42EP3 | CDC42EP3 | 4032 | 0.110 | 0.0213 | No |
| 26 | PLEKHF1 | PLEKHF1 | 4218 | 0.102 | 0.0164 | No |
| 27 | CD46 | CD46 | 4224 | 0.102 | 0.0190 | No |
| 28 | FUT9 | FUT9 | 4325 | 0.097 | 0.0174 | No |
| 29 | GLRX | GLRX | 4365 | 0.095 | 0.0184 | No |
| 30 | ETHE1 | ETHE1 | 4444 | 0.091 | 0.0176 | No |
| 31 | SMAGP | SMAGP | 4461 | 0.090 | 0.0194 | No |
| 32 | TSPAN7 | TSPAN7 | 4624 | 0.083 | 0.0150 | No |
| 33 | STC1 | STC1 | 4777 | 0.079 | 0.0108 | No |
| 34 | RHOG | RHOG | 5042 | 0.071 | 0.0018 | No |
| 35 | CHD7 | CHD7 | 5075 | 0.070 | 0.0024 | No |
| 36 | MVP | MVP | 5446 | 0.061 | -0.0112 | No |
| 37 | MYD88 | MYD88 | 5585 | 0.058 | -0.0154 | No |
| 38 | ARSA | ARSA | 5594 | 0.058 | -0.0141 | No |
| 39 | CTNND2 | CTNND2 | 5701 | 0.056 | -0.0170 | No |
| 40 | RNFT2 | RNFT2 | 6045 | 0.050 | -0.0298 | No |
| 41 | CEBPB | CEBPB | 6185 | 0.048 | -0.0343 | No |
| 42 | MAP2 | MAP2 | 6412 | 0.044 | -0.0425 | No |
| 43 | REXO2 | REXO2 | 6532 | 0.042 | -0.0462 | No |
| 44 | ATOX1 | ATOX1 | 6644 | 0.040 | -0.0497 | No |
| 45 | TSPAN1 | TSPAN1 | 6705 | 0.040 | -0.0512 | No |
| 46 | FADD | FADD | 6742 | 0.039 | -0.0516 | No |
| 47 | MYLK | MYLK | 6812 | 0.038 | -0.0534 | No |
| 48 | ITGA5 | ITGA5 | 6940 | 0.036 | -0.0577 | No |
| 49 | CLIP4 | CLIP4 | 7108 | 0.034 | -0.0637 | No |
| 50 | EDEM1 | EDEM1 | 7155 | 0.034 | -0.0646 | No |
| 51 | BET1L | BET1L | 7185 | 0.033 | -0.0649 | No |
| 52 | MARCKSL1 | MARCKSL1 | 7231 | 0.033 | -0.0659 | No |
| 53 | TMED9 | TMED9 | 7526 | 0.029 | -0.0773 | No |
| 54 | ANP32A | ANP32A | 7527 | 0.029 | -0.0765 | No |
| 55 | KCNAB2 | KCNAB2 | 7646 | 0.028 | -0.0806 | No |
| 56 | TPD52L2 | TPD52L2 | 7966 | 0.024 | -0.0932 | No |
| 57 | TCF4 | TCF4 | 8169 | 0.022 | -0.1009 | No |
| 58 | ARFRP1 | ARFRP1 | 8385 | 0.020 | -0.1093 | No |
| 59 | IFRD2 | IFRD2 | 8650 | 0.017 | -0.1197 | No |
| 60 | SSH3 | SSH3 | 8848 | 0.016 | -0.1274 | No |
| 61 | S100A10 | S100A10 | 8879 | 0.015 | -0.1283 | No |
| 62 | GEM | GEM | 8922 | 0.015 | -0.1296 | No |
| 63 | FOXC1 | FOXC1 | 8940 | 0.015 | -0.1299 | No |
| 64 | VASP | VASP | 9049 | 0.014 | -0.1340 | No |
| 65 | YIF1A | YIF1A | 9151 | 0.013 | -0.1378 | No |
| 66 | CDKN1B | CDKN1B | 9251 | 0.012 | -0.1416 | No |
| 67 | ABCD1 | ABCD1 | 9993 | 0.005 | -0.1721 | No |
| 68 | SLC38A10 | SLC38A10 | 10061 | 0.005 | -0.1747 | No |
| 69 | PIGT | PIGT | 10106 | 0.005 | -0.1764 | No |
| 70 | SLC4A7 | SLC4A7 | 10172 | 0.004 | -0.1790 | No |
| 71 | ANXA1 | ANXA1 | 10276 | 0.003 | -0.1832 | No |
| 72 | WDR55 | WDR55 | 10348 | 0.002 | -0.1861 | No |
| 73 | SLC35C1 | SLC35C1 | 14908 | -0.001 | -0.3746 | No |
| 74 | PRNP | PRNP | 15108 | -0.003 | -0.3827 | No |
| 75 | TMEM214 | TMEM214 | 15119 | -0.003 | -0.3831 | No |
| 76 | GSDMD | GSDMD | 15450 | -0.005 | -0.3966 | No |
| 77 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.4032 | No |
| 78 | ITGA6 | ITGA6 | 15693 | -0.008 | -0.4062 | No |
| 79 | FYCO1 | FYCO1 | 15713 | -0.008 | -0.4067 | No |
| 80 | SERPINE1 | SERPINE1 | 15830 | -0.009 | -0.4113 | No |
| 81 | DCBLD2 | DCBLD2 | 15941 | -0.010 | -0.4156 | No |
| 82 | MGAT4B | MGAT4B | 16035 | -0.011 | -0.4191 | No |
| 83 | FBRS | FBRS | 16041 | -0.011 | -0.4190 | No |
| 84 | RHBDF2 | RHBDF2 | 16132 | -0.012 | -0.4224 | No |
| 85 | ADRM1 | ADRM1 | 16232 | -0.013 | -0.4262 | No |
| 86 | S100A13 | S100A13 | 16435 | -0.015 | -0.4341 | No |
| 87 | DNAJC10 | DNAJC10 | 16505 | -0.016 | -0.4366 | No |
| 88 | KDELR3 | KDELR3 | 16508 | -0.016 | -0.4362 | No |
| 89 | DRAP1 | DRAP1 | 16540 | -0.016 | -0.4371 | No |
| 90 | RRBP1 | RRBP1 | 16591 | -0.016 | -0.4387 | No |
| 91 | COQ10B | COQ10B | 16755 | -0.018 | -0.4449 | No |
| 92 | EMP3 | EMP3 | 16762 | -0.018 | -0.4447 | No |
| 93 | TMEM109 | TMEM109 | 16979 | -0.021 | -0.4531 | No |
| 94 | QSOX1 | QSOX1 | 16995 | -0.021 | -0.4531 | No |
| 95 | DSTN | DSTN | 17009 | -0.021 | -0.4531 | No |
| 96 | FLII | FLII | 17011 | -0.021 | -0.4525 | No |
| 97 | ZMYM4 | ZMYM4 | 17046 | -0.022 | -0.4534 | No |
| 98 | EXOC3 | EXOC3 | 17088 | -0.022 | -0.4545 | No |
| 99 | DEGS1 | DEGS1 | 17089 | -0.022 | -0.4539 | No |
| 100 | B3GNT2 | B3GNT2 | 17124 | -0.023 | -0.4547 | No |
| 101 | YIPF3 | YIPF3 | 17160 | -0.023 | -0.4555 | No |
| 102 | MAFG | MAFG | 17175 | -0.023 | -0.4554 | No |
| 103 | LRRFIP2 | LRRFIP2 | 17343 | -0.025 | -0.4617 | No |
| 104 | ADSS | ADSS | 17435 | -0.026 | -0.4647 | No |
| 105 | PNPLA2 | PNPLA2 | 17451 | -0.026 | -0.4646 | No |
| 106 | HSPA5 | HSPA5 | 17529 | -0.027 | -0.4671 | No |
| 107 | PYGB | PYGB | 17535 | -0.028 | -0.4665 | No |
| 108 | CSRP1 | CSRP1 | 17567 | -0.028 | -0.4670 | No |
| 109 | MGAT1 | MGAT1 | 17682 | -0.030 | -0.4710 | No |
| 110 | NSMAF | NSMAF | 17710 | -0.030 | -0.4713 | No |
| 111 | LGALS8 | LGALS8 | 17818 | -0.032 | -0.4748 | No |
| 112 | CUL7 | CUL7 | 17865 | -0.032 | -0.4758 | No |
| 113 | MCFD2 | MCFD2 | 17913 | -0.033 | -0.4769 | No |
| 114 | PARVB | PARVB | 18018 | -0.035 | -0.4803 | No |
| 115 | CLIC1 | CLIC1 | 18140 | -0.037 | -0.4843 | No |
| 116 | GBE1 | GBE1 | 18170 | -0.037 | -0.4845 | No |
| 117 | GLIPR1 | GLIPR1 | 18201 | -0.038 | -0.4847 | No |
| 118 | SH3BGRL3 | SH3BGRL3 | 18251 | -0.039 | -0.4856 | No |
| 119 | ECM1 | ECM1 | 18376 | -0.041 | -0.4897 | No |
| 120 | IQGAP1 | IQGAP1 | 18498 | -0.043 | -0.4935 | No |
| 121 | CAPN2 | CAPN2 | 18601 | -0.046 | -0.4965 | No |
| 122 | FLNA | FLNA | 18719 | -0.048 | -0.5000 | No |
| 123 | LGALS1 | LGALS1 | 18926 | -0.053 | -0.5071 | No |
| 124 | FOXN3 | FOXN3 | 19085 | -0.058 | -0.5120 | No |
| 125 | PMAIP1 | PMAIP1 | 19107 | -0.058 | -0.5113 | No |
| 126 | FOSL2 | FOSL2 | 19180 | -0.061 | -0.5127 | No |
| 127 | PLCB1 | PLCB1 | 19196 | -0.061 | -0.5116 | No |
| 128 | ACTN4 | ACTN4 | 19250 | -0.063 | -0.5121 | No |
| 129 | TRAM2 | TRAM2 | 19266 | -0.064 | -0.5110 | No |
| 130 | SERINC3 | SERINC3 | 19317 | -0.065 | -0.5113 | No |
| 131 | PARVA | PARVA | 19372 | -0.067 | -0.5117 | No |
| 132 | CAV2 | CAV2 | 19408 | -0.069 | -0.5113 | No |
| 133 | VCL | VCL | 19438 | -0.070 | -0.5106 | No |
| 134 | PXN | PXN | 19583 | -0.075 | -0.5145 | No |
| 135 | IMPAD1 | IMPAD1 | 19595 | -0.075 | -0.5130 | No |
| 136 | KLHL21 | KLHL21 | 19631 | -0.077 | -0.5123 | No |
| 137 | FURIN | FURIN | 19684 | -0.079 | -0.5124 | No |
| 138 | TAPBP | TAPBP | 19699 | -0.079 | -0.5108 | No |
| 139 | ATP1A1 | ATP1A1 | 19740 | -0.081 | -0.5102 | No |
| 140 | WDR1 | WDR1 | 19790 | -0.084 | -0.5100 | No |
| 141 | PLP2 | PLP2 | 19799 | -0.084 | -0.5081 | No |
| 142 | LPP | LPP | 19943 | -0.091 | -0.5115 | No |
| 143 | CD44 | CD44 | 19954 | -0.091 | -0.5095 | No |
| 144 | ZBTB38 | ZBTB38 | 19975 | -0.092 | -0.5078 | No |
| 145 | SQSTM1 | SQSTM1 | 20077 | -0.098 | -0.5093 | No |
| 146 | IER3 | IER3 | 20182 | -0.102 | -0.5109 | No |
| 147 | ANXA4 | ANXA4 | 20186 | -0.103 | -0.5082 | No |
| 148 | RBKS | RBKS | 20507 | -0.117 | -0.5183 | Yes |
| 149 | SYNJ2 | SYNJ2 | 20519 | -0.118 | -0.5155 | Yes |
| 150 | ESYT1 | ESYT1 | 20568 | -0.120 | -0.5143 | Yes |
| 151 | MYH9 | MYH9 | 20636 | -0.124 | -0.5137 | Yes |
| 152 | RPS6KA4 | RPS6KA4 | 20745 | -0.130 | -0.5146 | Yes |
| 153 | BAG3 | BAG3 | 20851 | -0.138 | -0.5152 | Yes |
| 154 | SDC4 | SDC4 | 20885 | -0.139 | -0.5128 | Yes |
| 155 | RNH1 | RNH1 | 20892 | -0.140 | -0.5093 | Yes |
| 156 | MYO1C | MYO1C | 20897 | -0.140 | -0.5056 | Yes |
| 157 | GSTA4 | GSTA4 | 20940 | -0.145 | -0.5035 | Yes |
| 158 | KCND1 | KCND1 | 20941 | -0.145 | -0.4995 | Yes |
| 159 | CRIM1 | CRIM1 | 20947 | -0.146 | -0.4958 | Yes |
| 160 | UPP1 | UPP1 | 21058 | -0.157 | -0.4961 | Yes |
| 161 | EMP1 | EMP1 | 21196 | -0.170 | -0.4971 | Yes |
| 162 | FAH | FAH | 21237 | -0.174 | -0.4941 | Yes |
| 163 | CAST | CAST | 21363 | -0.190 | -0.4941 | Yes |
| 164 | PARP12 | PARP12 | 21383 | -0.192 | -0.4897 | Yes |
| 165 | CRTAM | CRTAM | 21490 | -0.203 | -0.4886 | Yes |
| 166 | LMNA | LMNA | 21572 | -0.210 | -0.4863 | Yes |
| 167 | ANXA11 | ANXA11 | 21579 | -0.211 | -0.4808 | Yes |
| 168 | SH2B3 | SH2B3 | 21658 | -0.219 | -0.4781 | Yes |
| 169 | SHC1 | SHC1 | 21741 | -0.230 | -0.4753 | Yes |
| 170 | SLC10A3 | SLC10A3 | 21813 | -0.241 | -0.4717 | Yes |
| 171 | RAVER2 | RAVER2 | 21909 | -0.255 | -0.4687 | Yes |
| 172 | ACTN1 | ACTN1 | 21926 | -0.257 | -0.4624 | Yes |
| 173 | BACE2 | BACE2 | 22035 | -0.275 | -0.4595 | Yes |
| 174 | P4HA2 | P4HA2 | 22070 | -0.281 | -0.4533 | Yes |
| 175 | NT5E | NT5E | 22108 | -0.287 | -0.4470 | Yes |
| 176 | COL6A2 | COL6A2 | 22117 | -0.287 | -0.4396 | Yes |
| 177 | RIN1 | RIN1 | 22120 | -0.288 | -0.4319 | Yes |
| 178 | ANPEP | ANPEP | 22140 | -0.292 | -0.4248 | Yes |
| 179 | TNFRSF12A | TNFRSF12A | 22148 | -0.293 | -0.4172 | Yes |
| 180 | PRSS23 | PRSS23 | 22159 | -0.295 | -0.4096 | Yes |
| 181 | RAC2 | RAC2 | 22161 | -0.295 | -0.4017 | Yes |
| 182 | PROCR | PROCR | 22311 | -0.323 | -0.3991 | Yes |
| 183 | PTGER4 | PTGER4 | 22332 | -0.329 | -0.3910 | Yes |
| 184 | COL18A1 | COL18A1 | 22357 | -0.332 | -0.3830 | Yes |
| 185 | ANXA2 | ANXA2 | 22374 | -0.334 | -0.3746 | Yes |
| 186 | IL11 | IL11 | 23152 | -0.395 | -0.3961 | Yes |
| 187 | PLAUR | PLAUR | 23251 | -0.408 | -0.3891 | Yes |
| 188 | AXL | AXL | 23290 | -0.416 | -0.3794 | Yes |
| 189 | PHLDA2 | PHLDA2 | 23371 | -0.437 | -0.3709 | Yes |
| 190 | PERP | PERP | 23434 | -0.459 | -0.3610 | Yes |
| 191 | OLFML2B | OLFML2B | 23440 | -0.462 | -0.3487 | Yes |
| 192 | ITGA3 | ITGA3 | 23486 | -0.477 | -0.3377 | Yes |
| 193 | S100A11 | S100A11 | 23510 | -0.487 | -0.3255 | Yes |
| 194 | PQLC3 | PQLC3 | 23524 | -0.491 | -0.3127 | Yes |
| 195 | AHNAK | AHNAK | 23609 | -0.531 | -0.3018 | Yes |
| 196 | MICAL2 | MICAL2 | 23614 | -0.532 | -0.2876 | Yes |
| 197 | MYOF | MYOF | 23756 | -0.619 | -0.2767 | Yes |
| 198 | CYB561 | CYB561 | 23857 | -0.641 | -0.2634 | Yes |
| 199 | IL18R1 | IL18R1 | 23880 | -0.650 | -0.2468 | Yes |
| 200 | FADS3 | FADS3 | 23982 | -0.718 | -0.2315 | Yes |
| 201 | MET | MET | 24021 | -0.756 | -0.2126 | Yes |
| 202 | TPM2 | TPM2 | 24050 | -0.788 | -0.1925 | Yes |
| 203 | CAV1 | CAV1 | 24167 | -1.007 | -0.1700 | Yes |
| 204 | FHL2 | FHL2 | 24205 | -1.090 | -0.1421 | Yes |
| 205 | SEMA4F | SEMA4F | 24247 | -1.192 | -0.1115 | Yes |
| 206 | EPHA2 | EPHA2 | 24249 | -1.201 | -0.0791 | Yes |
| 207 | S100A6 | S100A6 | 24307 | -1.423 | -0.0429 | Yes |
| 208 | GLT8D2 | GLT8D2 | 24346 | -1.708 | 0.0017 | Yes |