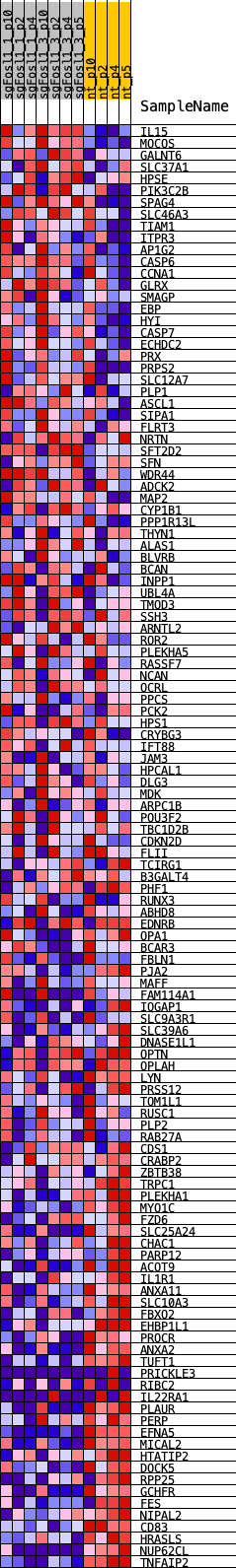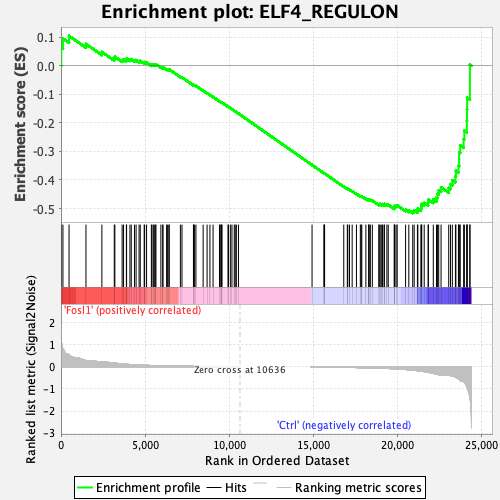
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | ELF4_REGULON |
| Enrichment Score (ES) | -0.51623195 |
| Normalized Enrichment Score (NES) | -1.4761231 |
| Nominal p-value | 0.023605151 |
| FDR q-value | 0.101584285 |
| FWER p-Value | 0.263 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IL15 | IL15 | 15 | 1.425 | 0.0632 | No |
| 2 | MOCOS | MOCOS | 115 | 0.810 | 0.0954 | No |
| 3 | GALNT6 | GALNT6 | 478 | 0.537 | 0.1046 | No |
| 4 | SLC37A1 | SLC37A1 | 1483 | 0.290 | 0.0762 | No |
| 5 | HPSE | HPSE | 2428 | 0.243 | 0.0482 | No |
| 6 | PIK3C2B | PIK3C2B | 3165 | 0.169 | 0.0255 | No |
| 7 | SPAG4 | SPAG4 | 3200 | 0.167 | 0.0316 | No |
| 8 | SLC46A3 | SLC46A3 | 3632 | 0.131 | 0.0197 | No |
| 9 | TIAM1 | TIAM1 | 3716 | 0.127 | 0.0220 | No |
| 10 | ITPR3 | ITPR3 | 3885 | 0.116 | 0.0202 | No |
| 11 | AP1G2 | AP1G2 | 3897 | 0.116 | 0.0250 | No |
| 12 | CASP6 | CASP6 | 4096 | 0.107 | 0.0216 | No |
| 13 | CCNA1 | CCNA1 | 4174 | 0.104 | 0.0231 | No |
| 14 | GLRX | GLRX | 4365 | 0.095 | 0.0195 | No |
| 15 | SMAGP | SMAGP | 4461 | 0.090 | 0.0196 | No |
| 16 | EBP | EBP | 4639 | 0.083 | 0.0160 | No |
| 17 | HYI | HYI | 4709 | 0.081 | 0.0168 | No |
| 18 | CASP7 | CASP7 | 4942 | 0.074 | 0.0106 | No |
| 19 | ECHDC2 | ECHDC2 | 4970 | 0.073 | 0.0128 | No |
| 20 | PRX | PRX | 5084 | 0.070 | 0.0112 | No |
| 21 | PRPS2 | PRPS2 | 5368 | 0.063 | 0.0024 | No |
| 22 | SLC12A7 | SLC12A7 | 5404 | 0.062 | 0.0037 | No |
| 23 | PLP1 | PLP1 | 5486 | 0.060 | 0.0031 | No |
| 24 | ASCL1 | ASCL1 | 5543 | 0.059 | 0.0034 | No |
| 25 | SIPA1 | SIPA1 | 5603 | 0.058 | 0.0036 | No |
| 26 | FLRT3 | FLRT3 | 5641 | 0.057 | 0.0046 | No |
| 27 | NRTN | NRTN | 5934 | 0.052 | -0.0051 | No |
| 28 | SFT2D2 | SFT2D2 | 6048 | 0.050 | -0.0075 | No |
| 29 | SFN | SFN | 6055 | 0.050 | -0.0055 | No |
| 30 | WDR44 | WDR44 | 6274 | 0.046 | -0.0125 | No |
| 31 | ADCK2 | ADCK2 | 6335 | 0.045 | -0.0129 | No |
| 32 | MAP2 | MAP2 | 6412 | 0.044 | -0.0141 | No |
| 33 | CYP1B1 | CYP1B1 | 6429 | 0.043 | -0.0128 | No |
| 34 | PPP1R13L | PPP1R13L | 7093 | 0.034 | -0.0386 | No |
| 35 | THYN1 | THYN1 | 7188 | 0.033 | -0.0410 | No |
| 36 | ALAS1 | ALAS1 | 7869 | 0.025 | -0.0679 | No |
| 37 | BLVRB | BLVRB | 7872 | 0.025 | -0.0668 | No |
| 38 | BCAN | BCAN | 7937 | 0.025 | -0.0683 | No |
| 39 | INPP1 | INPP1 | 8017 | 0.024 | -0.0705 | No |
| 40 | UBL4A | UBL4A | 8445 | 0.019 | -0.0873 | No |
| 41 | TMOD3 | TMOD3 | 8680 | 0.017 | -0.0961 | No |
| 42 | SSH3 | SSH3 | 8848 | 0.016 | -0.1023 | No |
| 43 | ARNTL2 | ARNTL2 | 9035 | 0.014 | -0.1094 | No |
| 44 | ROR2 | ROR2 | 9418 | 0.011 | -0.1246 | No |
| 45 | PLEKHA5 | PLEKHA5 | 9497 | 0.010 | -0.1274 | No |
| 46 | RASSF7 | RASSF7 | 9510 | 0.010 | -0.1275 | No |
| 47 | NCAN | NCAN | 9540 | 0.010 | -0.1282 | No |
| 48 | OCRL | OCRL | 9566 | 0.009 | -0.1288 | No |
| 49 | PPCS | PPCS | 9922 | 0.006 | -0.1432 | No |
| 50 | PCK2 | PCK2 | 9952 | 0.006 | -0.1441 | No |
| 51 | HPS1 | HPS1 | 10062 | 0.005 | -0.1484 | No |
| 52 | CRYBG3 | CRYBG3 | 10156 | 0.004 | -0.1521 | No |
| 53 | IFT88 | IFT88 | 10294 | 0.003 | -0.1576 | No |
| 54 | JAM3 | JAM3 | 10351 | 0.002 | -0.1598 | No |
| 55 | HPCAL1 | HPCAL1 | 10419 | 0.002 | -0.1625 | No |
| 56 | DLG3 | DLG3 | 10544 | 0.001 | -0.1675 | No |
| 57 | MDK | MDK | 14916 | -0.001 | -0.3476 | No |
| 58 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.3761 | No |
| 59 | POU3F2 | POU3F2 | 15656 | -0.007 | -0.3774 | No |
| 60 | TBC1D2B | TBC1D2B | 15661 | -0.007 | -0.3772 | No |
| 61 | CDKN2D | CDKN2D | 16800 | -0.019 | -0.4233 | No |
| 62 | FLII | FLII | 17011 | -0.021 | -0.4310 | No |
| 63 | TCIRG1 | TCIRG1 | 17040 | -0.021 | -0.4312 | No |
| 64 | B3GALT4 | B3GALT4 | 17140 | -0.023 | -0.4343 | No |
| 65 | PHF1 | PHF1 | 17299 | -0.025 | -0.4397 | No |
| 66 | RUNX3 | RUNX3 | 17554 | -0.028 | -0.4489 | No |
| 67 | ABHD8 | ABHD8 | 17784 | -0.031 | -0.4569 | No |
| 68 | EDNRB | EDNRB | 17833 | -0.032 | -0.4575 | No |
| 69 | OPA1 | OPA1 | 17872 | -0.032 | -0.4576 | No |
| 70 | BCAR3 | BCAR3 | 18113 | -0.036 | -0.4658 | No |
| 71 | FBLN1 | FBLN1 | 18281 | -0.040 | -0.4710 | No |
| 72 | PJA2 | PJA2 | 18298 | -0.040 | -0.4698 | No |
| 73 | MAFF | MAFF | 18299 | -0.040 | -0.4680 | No |
| 74 | FAM114A1 | FAM114A1 | 18380 | -0.041 | -0.4695 | No |
| 75 | IQGAP1 | IQGAP1 | 18498 | -0.043 | -0.4724 | No |
| 76 | SLC9A3R1 | SLC9A3R1 | 18878 | -0.052 | -0.4857 | No |
| 77 | SLC39A6 | SLC39A6 | 18941 | -0.054 | -0.4858 | No |
| 78 | DNASE1L1 | DNASE1L1 | 18958 | -0.054 | -0.4840 | No |
| 79 | OPTN | OPTN | 19056 | -0.057 | -0.4855 | No |
| 80 | OPLAH | OPLAH | 19104 | -0.058 | -0.4848 | No |
| 81 | LYN | LYN | 19195 | -0.061 | -0.4858 | No |
| 82 | PRSS12 | PRSS12 | 19214 | -0.062 | -0.4837 | No |
| 83 | TOM1L1 | TOM1L1 | 19363 | -0.067 | -0.4868 | No |
| 84 | RUSC1 | RUSC1 | 19443 | -0.070 | -0.4870 | No |
| 85 | PLP2 | PLP2 | 19799 | -0.084 | -0.4978 | No |
| 86 | RAB27A | RAB27A | 19809 | -0.084 | -0.4944 | No |
| 87 | CDS1 | CDS1 | 19828 | -0.085 | -0.4913 | No |
| 88 | CRABP2 | CRABP2 | 19907 | -0.089 | -0.4905 | No |
| 89 | ZBTB38 | ZBTB38 | 19975 | -0.092 | -0.4892 | No |
| 90 | TRPC1 | TRPC1 | 20473 | -0.116 | -0.5045 | No |
| 91 | PLEKHA1 | PLEKHA1 | 20664 | -0.125 | -0.5067 | No |
| 92 | MYO1C | MYO1C | 20897 | -0.140 | -0.5099 | Yes |
| 93 | FZD6 | FZD6 | 20984 | -0.150 | -0.5068 | Yes |
| 94 | SLC25A24 | SLC25A24 | 21176 | -0.168 | -0.5071 | Yes |
| 95 | CHAC1 | CHAC1 | 21202 | -0.170 | -0.5005 | Yes |
| 96 | PARP12 | PARP12 | 21383 | -0.192 | -0.4994 | Yes |
| 97 | ACOT9 | ACOT9 | 21414 | -0.195 | -0.4919 | Yes |
| 98 | IL1R1 | IL1R1 | 21433 | -0.197 | -0.4838 | Yes |
| 99 | ANXA11 | ANXA11 | 21579 | -0.211 | -0.4803 | Yes |
| 100 | SLC10A3 | SLC10A3 | 21813 | -0.241 | -0.4791 | Yes |
| 101 | FBXO2 | FBXO2 | 21827 | -0.243 | -0.4688 | Yes |
| 102 | EHBP1L1 | EHBP1L1 | 22119 | -0.288 | -0.4679 | Yes |
| 103 | PROCR | PROCR | 22311 | -0.323 | -0.4613 | Yes |
| 104 | ANXA2 | ANXA2 | 22374 | -0.334 | -0.4489 | Yes |
| 105 | TUFT1 | TUFT1 | 22441 | -0.348 | -0.4360 | Yes |
| 106 | PRICKLE3 | PRICKLE3 | 22581 | -0.352 | -0.4260 | Yes |
| 107 | RIBC2 | RIBC2 | 23036 | -0.370 | -0.4281 | Yes |
| 108 | IL22RA1 | IL22RA1 | 23141 | -0.393 | -0.4147 | Yes |
| 109 | PLAUR | PLAUR | 23251 | -0.408 | -0.4010 | Yes |
| 110 | PERP | PERP | 23434 | -0.459 | -0.3879 | Yes |
| 111 | EFNA5 | EFNA5 | 23462 | -0.471 | -0.3679 | Yes |
| 112 | MICAL2 | MICAL2 | 23614 | -0.532 | -0.3503 | Yes |
| 113 | HTATIP2 | HTATIP2 | 23654 | -0.546 | -0.3274 | Yes |
| 114 | DOCK5 | DOCK5 | 23656 | -0.548 | -0.3029 | Yes |
| 115 | RPP25 | RPP25 | 23719 | -0.588 | -0.2791 | Yes |
| 116 | GCHFR | GCHFR | 23924 | -0.672 | -0.2574 | Yes |
| 117 | FES | FES | 23965 | -0.703 | -0.2275 | Yes |
| 118 | NIPAL2 | NIPAL2 | 24113 | -0.895 | -0.1935 | Yes |
| 119 | CD83 | CD83 | 24121 | -0.915 | -0.1528 | Yes |
| 120 | HRASLS | HRASLS | 24130 | -0.922 | -0.1118 | Yes |
| 121 | NUP62CL | NUP62CL | 24294 | -1.361 | -0.0575 | Yes |
| 122 | TNFAIP2 | TNFAIP2 | 24295 | -1.370 | 0.0038 | Yes |