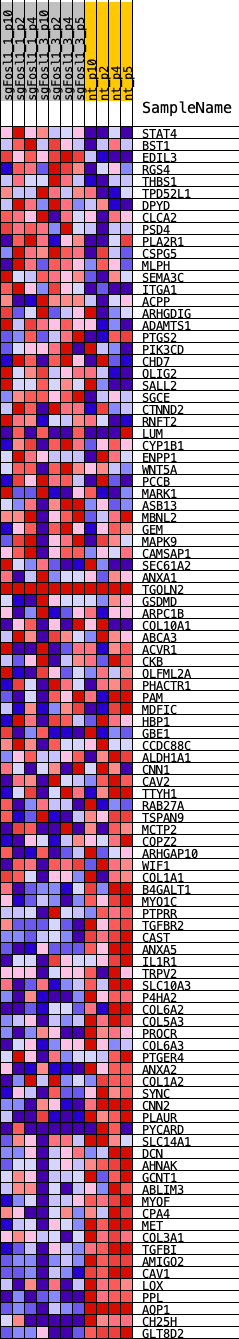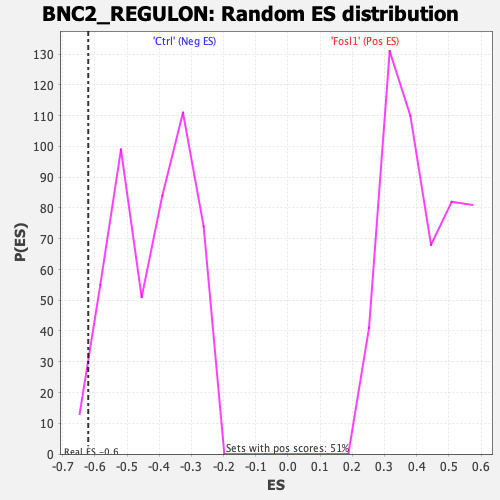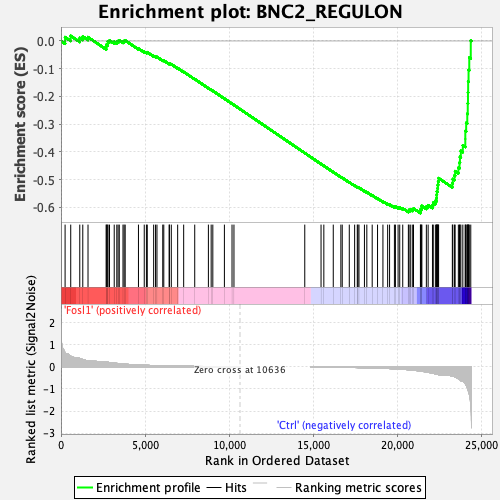
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NSCs_shNf1_sgFosl1.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl.NSCs_shNf1_sgFosl1.cls #Fosl1_versus_Ctrl_repos |
| Phenotype | NSCs_shNf1_sgFosl1.cls#Fosl1_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | BNC2_REGULON |
| Enrichment Score (ES) | -0.6206039 |
| Normalized Enrichment Score (NES) | -1.4796375 |
| Nominal p-value | 0.02053388 |
| FDR q-value | 0.1164419 |
| FWER p-Value | 0.243 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | STAT4 | STAT4 | 243 | 0.625 | 0.0139 | No |
| 2 | BST1 | BST1 | 578 | 0.480 | 0.0184 | No |
| 3 | EDIL3 | EDIL3 | 1115 | 0.382 | 0.0109 | No |
| 4 | RGS4 | RGS4 | 1294 | 0.331 | 0.0162 | No |
| 5 | THBS1 | THBS1 | 1605 | 0.272 | 0.0139 | No |
| 6 | TPD52L1 | TPD52L1 | 2689 | 0.223 | -0.0222 | No |
| 7 | DPYD | DPYD | 2692 | 0.223 | -0.0138 | No |
| 8 | CLCA2 | CLCA2 | 2778 | 0.210 | -0.0093 | No |
| 9 | PSD4 | PSD4 | 2785 | 0.209 | -0.0015 | No |
| 10 | PLA2R1 | PLA2R1 | 2882 | 0.197 | 0.0021 | No |
| 11 | CSPG5 | CSPG5 | 3162 | 0.170 | -0.0029 | No |
| 12 | MLPH | MLPH | 3316 | 0.158 | -0.0032 | No |
| 13 | SEMA3C | SEMA3C | 3390 | 0.150 | -0.0005 | No |
| 14 | ITGA1 | ITGA1 | 3462 | 0.143 | 0.0021 | No |
| 15 | ACPP | ACPP | 3683 | 0.129 | -0.0020 | No |
| 16 | ARHGDIG | ARHGDIG | 3750 | 0.124 | -0.0000 | No |
| 17 | ADAMTS1 | ADAMTS1 | 3813 | 0.120 | 0.0020 | No |
| 18 | PTGS2 | PTGS2 | 4602 | 0.084 | -0.0272 | No |
| 19 | PIK3CD | PIK3CD | 4946 | 0.074 | -0.0385 | No |
| 20 | CHD7 | CHD7 | 5075 | 0.070 | -0.0411 | No |
| 21 | OLIG2 | OLIG2 | 5125 | 0.069 | -0.0405 | No |
| 22 | SALL2 | SALL2 | 5505 | 0.060 | -0.0538 | No |
| 23 | SGCE | SGCE | 5618 | 0.058 | -0.0562 | No |
| 24 | CTNND2 | CTNND2 | 5701 | 0.056 | -0.0575 | No |
| 25 | RNFT2 | RNFT2 | 6045 | 0.050 | -0.0697 | No |
| 26 | LUM | LUM | 6107 | 0.049 | -0.0703 | No |
| 27 | CYP1B1 | CYP1B1 | 6429 | 0.043 | -0.0819 | No |
| 28 | ENPP1 | ENPP1 | 6438 | 0.043 | -0.0806 | No |
| 29 | WNT5A | WNT5A | 6553 | 0.042 | -0.0837 | No |
| 30 | PCCB | PCCB | 6931 | 0.036 | -0.0978 | No |
| 31 | MARK1 | MARK1 | 7288 | 0.032 | -0.1112 | No |
| 32 | ASB13 | ASB13 | 7945 | 0.025 | -0.1373 | No |
| 33 | MBNL2 | MBNL2 | 8757 | 0.016 | -0.1701 | No |
| 34 | GEM | GEM | 8922 | 0.015 | -0.1763 | No |
| 35 | MAPK9 | MAPK9 | 9020 | 0.014 | -0.1797 | No |
| 36 | CAMSAP1 | CAMSAP1 | 9708 | 0.008 | -0.2077 | No |
| 37 | SEC61A2 | SEC61A2 | 10161 | 0.004 | -0.2262 | No |
| 38 | ANXA1 | ANXA1 | 10276 | 0.003 | -0.2307 | No |
| 39 | TGOLN2 | TGOLN2 | 14483 | 0.000 | -0.4039 | No |
| 40 | GSDMD | GSDMD | 15450 | -0.005 | -0.4435 | No |
| 41 | ARPC1B | ARPC1B | 15616 | -0.007 | -0.4500 | No |
| 42 | COL10A1 | COL10A1 | 16176 | -0.012 | -0.4725 | No |
| 43 | ABCA3 | ABCA3 | 16628 | -0.017 | -0.4905 | No |
| 44 | ACVR1 | ACVR1 | 16709 | -0.018 | -0.4931 | No |
| 45 | CKB | CKB | 17128 | -0.023 | -0.5094 | No |
| 46 | OLFML2A | OLFML2A | 17446 | -0.026 | -0.5215 | No |
| 47 | PHACTR1 | PHACTR1 | 17603 | -0.028 | -0.5268 | No |
| 48 | PAM | PAM | 17642 | -0.029 | -0.5273 | No |
| 49 | MDFIC | MDFIC | 17704 | -0.030 | -0.5287 | No |
| 50 | HBP1 | HBP1 | 18028 | -0.035 | -0.5406 | No |
| 51 | GBE1 | GBE1 | 18170 | -0.037 | -0.5450 | No |
| 52 | CCDC88C | CCDC88C | 18489 | -0.043 | -0.5564 | No |
| 53 | ALDH1A1 | ALDH1A1 | 18807 | -0.050 | -0.5676 | No |
| 54 | CNN1 | CNN1 | 19127 | -0.059 | -0.5785 | No |
| 55 | CAV2 | CAV2 | 19408 | -0.069 | -0.5874 | No |
| 56 | TTYH1 | TTYH1 | 19522 | -0.072 | -0.5892 | No |
| 57 | RAB27A | RAB27A | 19809 | -0.084 | -0.5978 | No |
| 58 | TSPAN9 | TSPAN9 | 19869 | -0.088 | -0.5969 | No |
| 59 | MCTP2 | MCTP2 | 20028 | -0.094 | -0.5998 | No |
| 60 | COPZ2 | COPZ2 | 20125 | -0.100 | -0.5999 | No |
| 61 | ARHGAP10 | ARHGAP10 | 20304 | -0.107 | -0.6032 | No |
| 62 | WIF1 | WIF1 | 20638 | -0.124 | -0.6121 | No |
| 63 | COL1A1 | COL1A1 | 20681 | -0.127 | -0.6090 | No |
| 64 | B4GALT1 | B4GALT1 | 20779 | -0.132 | -0.6080 | No |
| 65 | MYO1C | MYO1C | 20897 | -0.140 | -0.6074 | No |
| 66 | PTPRR | PTPRR | 20943 | -0.145 | -0.6037 | No |
| 67 | TGFBR2 | TGFBR2 | 21354 | -0.188 | -0.6134 | Yes |
| 68 | CAST | CAST | 21363 | -0.190 | -0.6065 | Yes |
| 69 | ANXA5 | ANXA5 | 21429 | -0.196 | -0.6017 | Yes |
| 70 | IL1R1 | IL1R1 | 21433 | -0.197 | -0.5943 | Yes |
| 71 | TRPV2 | TRPV2 | 21707 | -0.225 | -0.5970 | Yes |
| 72 | SLC10A3 | SLC10A3 | 21813 | -0.241 | -0.5921 | Yes |
| 73 | P4HA2 | P4HA2 | 22070 | -0.281 | -0.5919 | Yes |
| 74 | COL6A2 | COL6A2 | 22117 | -0.287 | -0.5829 | Yes |
| 75 | COL5A3 | COL5A3 | 22243 | -0.310 | -0.5762 | Yes |
| 76 | PROCR | PROCR | 22311 | -0.323 | -0.5666 | Yes |
| 77 | COL6A3 | COL6A3 | 22321 | -0.326 | -0.5545 | Yes |
| 78 | PTGER4 | PTGER4 | 22332 | -0.329 | -0.5424 | Yes |
| 79 | ANXA2 | ANXA2 | 22374 | -0.334 | -0.5313 | Yes |
| 80 | COL1A2 | COL1A2 | 22387 | -0.337 | -0.5189 | Yes |
| 81 | SYNC | SYNC | 22408 | -0.343 | -0.5066 | Yes |
| 82 | CNN2 | CNN2 | 22435 | -0.347 | -0.4944 | Yes |
| 83 | PLAUR | PLAUR | 23251 | -0.408 | -0.5124 | Yes |
| 84 | PYCARD | PYCARD | 23277 | -0.414 | -0.4977 | Yes |
| 85 | SLC14A1 | SLC14A1 | 23375 | -0.438 | -0.4849 | Yes |
| 86 | DCN | DCN | 23416 | -0.453 | -0.4693 | Yes |
| 87 | AHNAK | AHNAK | 23609 | -0.531 | -0.4569 | Yes |
| 88 | GCNT1 | GCNT1 | 23676 | -0.557 | -0.4384 | Yes |
| 89 | ABLIM3 | ABLIM3 | 23703 | -0.575 | -0.4175 | Yes |
| 90 | MYOF | MYOF | 23756 | -0.619 | -0.3960 | Yes |
| 91 | CPA4 | CPA4 | 23874 | -0.646 | -0.3762 | Yes |
| 92 | MET | MET | 24021 | -0.756 | -0.3533 | Yes |
| 93 | COL3A1 | COL3A1 | 24022 | -0.757 | -0.3245 | Yes |
| 94 | TGFBI | TGFBI | 24073 | -0.818 | -0.2953 | Yes |
| 95 | AMIGO2 | AMIGO2 | 24139 | -0.934 | -0.2623 | Yes |
| 96 | CAV1 | CAV1 | 24167 | -1.007 | -0.2250 | Yes |
| 97 | LOX | LOX | 24181 | -1.034 | -0.1860 | Yes |
| 98 | PPL | PPL | 24191 | -1.057 | -0.1461 | Yes |
| 99 | AQP1 | AQP1 | 24215 | -1.107 | -0.1048 | Yes |
| 100 | CH25H | CH25H | 24257 | -1.221 | -0.0599 | Yes |
| 101 | GLT8D2 | GLT8D2 | 24346 | -1.708 | 0.0017 | Yes |