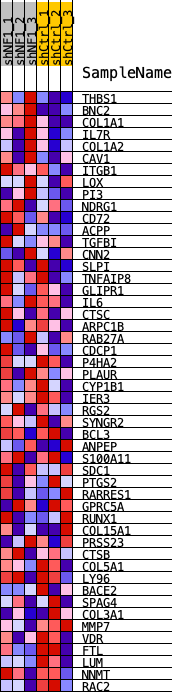
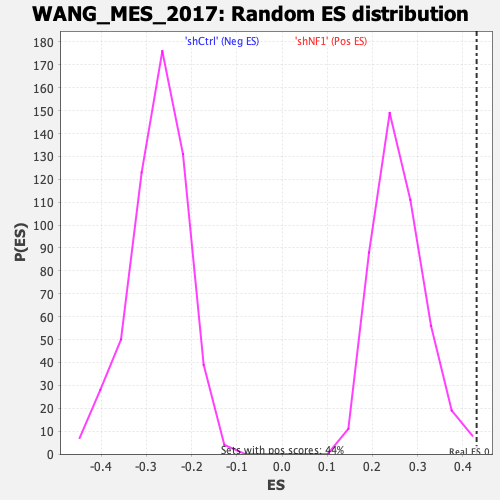
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shNF1 |
| GeneSet | WANG_MES_2017 |
| Enrichment Score (ES) | 0.4302045 |
| Normalized Enrichment Score (NES) | 1.6611276 |
| Nominal p-value | 0.009049774 |
| FDR q-value | 0.004547161 |
| FWER p-Value | 0.03 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | THBS1 | THBS1 | 13 | 0.123 | 0.0972 | Yes |
| 2 | BNC2 | BNC2 | 40 | 0.102 | 0.1769 | Yes |
| 3 | COL1A1 | COL1A1 | 158 | 0.082 | 0.2365 | Yes |
| 4 | IL7R | IL7R | 279 | 0.072 | 0.2878 | Yes |
| 5 | COL1A2 | COL1A2 | 651 | 0.057 | 0.3160 | Yes |
| 6 | CAV1 | CAV1 | 814 | 0.054 | 0.3514 | Yes |
| 7 | ITGB1 | ITGB1 | 957 | 0.051 | 0.3854 | Yes |
| 8 | LOX | LOX | 1391 | 0.045 | 0.4007 | Yes |
| 9 | PI3 | PI3 | 1922 | 0.039 | 0.4073 | Yes |
| 10 | NDRG1 | NDRG1 | 2507 | 0.034 | 0.4074 | Yes |
| 11 | CD72 | CD72 | 2847 | 0.032 | 0.4170 | Yes |
| 12 | ACPP | ACPP | 3084 | 0.030 | 0.4302 | Yes |
| 13 | TGFBI | TGFBI | 4792 | 0.021 | 0.3678 | No |
| 14 | CNN2 | CNN2 | 5091 | 0.020 | 0.3696 | No |
| 15 | SLPI | SLPI | 5413 | 0.018 | 0.3693 | No |
| 16 | TNFAIP8 | TNFAIP8 | 5465 | 0.018 | 0.3813 | No |
| 17 | GLIPR1 | GLIPR1 | 5605 | 0.017 | 0.3887 | No |
| 18 | IL6 | IL6 | 5609 | 0.017 | 0.4023 | No |
| 19 | CTSC | CTSC | 6083 | 0.016 | 0.3927 | No |
| 20 | ARPC1B | ARPC1B | 6192 | 0.015 | 0.3997 | No |
| 21 | RAB27A | RAB27A | 6204 | 0.015 | 0.4111 | No |
| 22 | CDCP1 | CDCP1 | 6238 | 0.015 | 0.4213 | No |
| 23 | P4HA2 | P4HA2 | 7058 | 0.012 | 0.3926 | No |
| 24 | PLAUR | PLAUR | 7477 | 0.010 | 0.3814 | No |
| 25 | CYP1B1 | CYP1B1 | 7673 | 0.010 | 0.3801 | No |
| 26 | IER3 | IER3 | 8602 | 0.007 | 0.3422 | No |
| 27 | RGS2 | RGS2 | 9049 | 0.005 | 0.3255 | No |
| 28 | SYNGR2 | SYNGR2 | 9437 | 0.004 | 0.3107 | No |
| 29 | BCL3 | BCL3 | 9852 | 0.002 | 0.2934 | No |
| 30 | ANPEP | ANPEP | 9908 | 0.002 | 0.2926 | No |
| 31 | S100A11 | S100A11 | 10703 | -0.000 | 0.2559 | No |
| 32 | SDC1 | SDC1 | 11233 | -0.002 | 0.2327 | No |
| 33 | PTGS2 | PTGS2 | 11356 | -0.002 | 0.2289 | No |
| 34 | RARRES1 | RARRES1 | 11546 | -0.003 | 0.2224 | No |
| 35 | GPRC5A | GPRC5A | 11746 | -0.004 | 0.2159 | No |
| 36 | RUNX1 | RUNX1 | 12042 | -0.005 | 0.2059 | No |
| 37 | COL15A1 | COL15A1 | 12302 | -0.005 | 0.1981 | No |
| 38 | PRSS23 | PRSS23 | 12359 | -0.006 | 0.2000 | No |
| 39 | CTSB | CTSB | 12661 | -0.007 | 0.1912 | No |
| 40 | COL5A1 | COL5A1 | 14074 | -0.011 | 0.1346 | No |
| 41 | LY96 | LY96 | 14269 | -0.012 | 0.1351 | No |
| 42 | BACE2 | BACE2 | 15352 | -0.016 | 0.0977 | No |
| 43 | SPAG4 | SPAG4 | 16140 | -0.020 | 0.0768 | No |
| 44 | COL3A1 | COL3A1 | 16349 | -0.021 | 0.0834 | No |
| 45 | MMP7 | MMP7 | 16754 | -0.023 | 0.0825 | No |
| 46 | VDR | VDR | 17575 | -0.027 | 0.0656 | No |
| 47 | FTL | FTL | 17640 | -0.027 | 0.0841 | No |
| 48 | LUM | LUM | 18137 | -0.030 | 0.0848 | No |
| 49 | NNMT | NNMT | 18514 | -0.032 | 0.0929 | No |
| 50 | RAC2 | RAC2 | 21019 | -0.063 | 0.0268 | No |