Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | VERHAAK_PN_2010 |
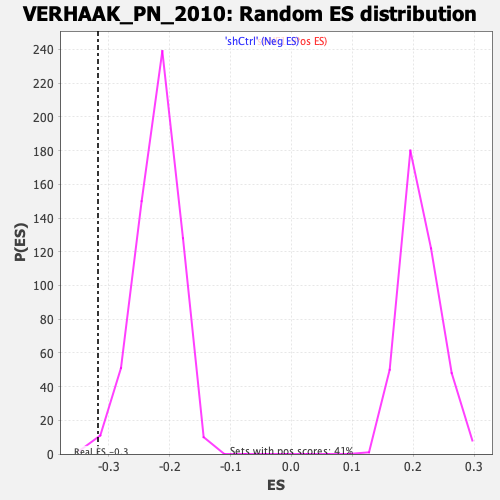
| Enrichment Score (ES) | -0.31741506 |
| Normalized Enrichment Score (NES) | -1.4421126 |
| Nominal p-value | 0.005076142 |
| FDR q-value | 0.03953936 |
| FWER p-Value | 0.315 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ALCAM | ALCAM | 45 | 0.100 | 0.0191 | No |
| 2 | CBX1 | CBX1 | 58 | 0.098 | 0.0392 | No |
| 3 | RAP2A | RAP2A | 236 | 0.075 | 0.0468 | No |
| 4 | ZNF286A | ZNF286A | 295 | 0.070 | 0.0589 | No |
| 5 | GPM6A | GPM6A | 422 | 0.065 | 0.0667 | No |
| 6 | CNTN1 | CNTN1 | 452 | 0.063 | 0.0787 | No |
| 7 | DNM3 | DNM3 | 548 | 0.060 | 0.0869 | No |
| 8 | ACTR1A | ACTR1A | 747 | 0.055 | 0.0894 | No |
| 9 | TOP2B | TOP2B | 794 | 0.055 | 0.0987 | No |
| 10 | CAMSAP1L1 | CAMSAP1L1 | 829 | 0.054 | 0.1085 | No |
| 11 | DBN1 | DBN1 | 840 | 0.054 | 0.1194 | No |
| 12 | RAD21 | RAD21 | 863 | 0.053 | 0.1296 | No |
| 13 | GABRA3 | GABRA3 | 989 | 0.051 | 0.1344 | No |
| 14 | FHOD3 | FHOD3 | 1516 | 0.043 | 0.1190 | No |
| 15 | ZNF510 | ZNF510 | 1794 | 0.041 | 0.1146 | No |
| 16 | BCOR | BCOR | 1830 | 0.040 | 0.1215 | No |
| 17 | SEZ6L | SEZ6L | 2022 | 0.038 | 0.1207 | No |
| 18 | TTC3 | TTC3 | 2176 | 0.037 | 0.1213 | No |
| 19 | HDAC2 | HDAC2 | 2530 | 0.034 | 0.1121 | No |
| 20 | NOL4 | NOL4 | 3041 | 0.031 | 0.0948 | No |
| 21 | LPHN3 | LPHN3 | 3072 | 0.031 | 0.0998 | No |
| 22 | E2F3 | E2F3 | 3108 | 0.030 | 0.1046 | No |
| 23 | HOXD3 | HOXD3 | 3432 | 0.028 | 0.0955 | No |
| 24 | NRXN2 | NRXN2 | 3492 | 0.028 | 0.0986 | No |
| 25 | SOX11 | SOX11 | 3515 | 0.028 | 0.1034 | No |
| 26 | FGF9 | FGF9 | 3578 | 0.027 | 0.1063 | No |
| 27 | SLC1A1 | SLC1A1 | 3612 | 0.027 | 0.1105 | No |
| 28 | PAK3 | PAK3 | 3667 | 0.027 | 0.1136 | No |
| 29 | MLLT11 | MLLT11 | 4075 | 0.025 | 0.0998 | No |
| 30 | BCL7A | BCL7A | 4126 | 0.024 | 0.1026 | No |
| 31 | SLCO5A1 | SLCO5A1 | 4299 | 0.024 | 0.0995 | No |
| 32 | ZNF184 | ZNF184 | 4498 | 0.023 | 0.0951 | No |
| 33 | MPPED2 | MPPED2 | 4550 | 0.022 | 0.0974 | No |
| 34 | ZEB2 | ZEB2 | 4614 | 0.022 | 0.0991 | No |
| 35 | TNRC4 | TNRC4 | 5083 | 0.020 | 0.0814 | No |
| 36 | DPP6 | DPP6 | 5104 | 0.020 | 0.0847 | No |
| 37 | CLASP2 | CLASP2 | 5119 | 0.020 | 0.0882 | No |
| 38 | PAK7 | PAK7 | 5215 | 0.019 | 0.0878 | No |
| 39 | PCDH11Y | PCDH11Y | 5265 | 0.019 | 0.0896 | No |
| 40 | MMP16 | MMP16 | 5348 | 0.019 | 0.0897 | No |
| 41 | VEZF1 | VEZF1 | 5397 | 0.018 | 0.0913 | No |
| 42 | DCX | DCX | 5806 | 0.017 | 0.0758 | No |
| 43 | NCAM1 | NCAM1 | 6426 | 0.014 | 0.0499 | No |
| 44 | LRRTM4 | LRRTM4 | 6429 | 0.014 | 0.0528 | No |
| 45 | ATAD5 | ATAD5 | 6435 | 0.014 | 0.0555 | No |
| 46 | TOPBP1 | TOPBP1 | 6531 | 0.014 | 0.0540 | No |
| 47 | GRID2 | GRID2 | 6874 | 0.012 | 0.0406 | No |
| 48 | SPTBN2 | SPTBN2 | 7001 | 0.012 | 0.0373 | No |
| 49 | CXXC4 | CXXC4 | 7355 | 0.011 | 0.0230 | No |
| 50 | KLRC4 | KLRC4 | 7526 | 0.010 | 0.0173 | No |
| 51 | FBXO21 | FBXO21 | 7728 | 0.010 | 0.0099 | No |
| 52 | SRGAP3 | SRGAP3 | 7746 | 0.009 | 0.0111 | No |
| 53 | GSK3B | GSK3B | 7802 | 0.009 | 0.0105 | No |
| 54 | MATR3 | MATR3 | 8083 | 0.008 | -0.0009 | No |
| 55 | FLRT1 | FLRT1 | 8146 | 0.008 | -0.0021 | No |
| 56 | ATP1A3 | ATP1A3 | 8391 | 0.007 | -0.0119 | No |
| 57 | CDKN1B | CDKN1B | 8492 | 0.007 | -0.0151 | No |
| 58 | TMEM35 | TMEM35 | 8601 | 0.007 | -0.0188 | No |
| 59 | KIF21B | KIF21B | 8787 | 0.006 | -0.0261 | No |
| 60 | EPB41 | EPB41 | 8844 | 0.006 | -0.0275 | No |
| 61 | ABAT | ABAT | 9125 | 0.005 | -0.0396 | No |
| 62 | KLRK1 | KLRK1 | 9147 | 0.005 | -0.0396 | No |
| 63 | C6orf134 | C6orf134 | 9264 | 0.004 | -0.0440 | No |
| 64 | ZNF248 | ZNF248 | 9318 | 0.004 | -0.0456 | No |
| 65 | PFN2 | PFN2 | 9445 | 0.004 | -0.0507 | No |
| 66 | PURG | PURG | 9522 | 0.004 | -0.0535 | No |
| 67 | SNX26 | SNX26 | 9529 | 0.004 | -0.0530 | No |
| 68 | MCM10 | MCM10 | 9565 | 0.003 | -0.0539 | No |
| 69 | CLGN | CLGN | 9585 | 0.003 | -0.0541 | No |
| 70 | SOX10 | SOX10 | 9761 | 0.003 | -0.0617 | No |
| 71 | ZNF711 | ZNF711 | 9786 | 0.003 | -0.0622 | No |
| 72 | CDC25A | CDC25A | 9788 | 0.003 | -0.0617 | No |
| 73 | CASK | CASK | 9890 | 0.002 | -0.0659 | No |
| 74 | RALGPS2 | RALGPS2 | 9934 | 0.002 | -0.0675 | No |
| 75 | CA10 | CA10 | 9972 | 0.002 | -0.0688 | No |
| 76 | CHD7 | CHD7 | 9974 | 0.002 | -0.0684 | No |
| 77 | AOF2 | AOF2 | 9990 | 0.002 | -0.0686 | No |
| 78 | KLRC3 | KLRC3 | 10008 | 0.002 | -0.0690 | No |
| 79 | STMN1 | STMN1 | 10027 | 0.002 | -0.0695 | No |
| 80 | MARCKS | MARCKS | 10036 | 0.002 | -0.0694 | No |
| 81 | WASF1 | WASF1 | 10041 | 0.002 | -0.0692 | No |
| 82 | SATB1 | SATB1 | 10095 | 0.002 | -0.0714 | No |
| 83 | DGKI | DGKI | 10108 | 0.002 | -0.0716 | No |
| 84 | RALGPS1 | RALGPS1 | 10203 | 0.001 | -0.0757 | No |
| 85 | TAF5 | TAF5 | 10419 | 0.001 | -0.0855 | No |
| 86 | FAM125B | FAM125B | 10490 | 0.001 | -0.0887 | No |
| 87 | RUFY3 | RUFY3 | 10556 | 0.000 | -0.0917 | No |
| 88 | PELI1 | PELI1 | 10697 | -0.000 | -0.0982 | No |
| 89 | TSPAN3 | TSPAN3 | 10699 | -0.000 | -0.0982 | No |
| 90 | MMP15 | MMP15 | 11307 | -0.002 | -0.1261 | No |
| 91 | IL1RAPL1 | IL1RAPL1 | 11319 | -0.002 | -0.1262 | No |
| 92 | CDC7 | CDC7 | 11332 | -0.002 | -0.1263 | No |
| 93 | FAM110B | FAM110B | 11490 | -0.003 | -0.1330 | No |
| 94 | PCDH11X | PCDH11X | 11637 | -0.003 | -0.1392 | No |
| 95 | NRXN1 | NRXN1 | 11758 | -0.004 | -0.1440 | No |
| 96 | ZNF643 | ZNF643 | 11822 | -0.004 | -0.1462 | No |
| 97 | MYO10 | MYO10 | 11837 | -0.004 | -0.1460 | No |
| 98 | TMCC1 | TMCC1 | 11903 | -0.004 | -0.1482 | No |
| 99 | MAPT | MAPT | 12192 | -0.005 | -0.1606 | No |
| 100 | SORCS3 | SORCS3 | 12363 | -0.006 | -0.1673 | No |
| 101 | PLCB4 | PLCB4 | 12472 | -0.006 | -0.1711 | No |
| 102 | P2RX7 | P2RX7 | 12774 | -0.007 | -0.1837 | No |
| 103 | STMN4 | STMN4 | 12976 | -0.008 | -0.1914 | No |
| 104 | PDE10A | PDE10A | 12988 | -0.008 | -0.1903 | No |
| 105 | MYB | MYB | 13495 | -0.009 | -0.2120 | No |
| 106 | SCN3A | SCN3A | 13980 | -0.011 | -0.2323 | No |
| 107 | MAP2 | MAP2 | 14012 | -0.011 | -0.2314 | No |
| 108 | LRP6 | LRP6 | 14070 | -0.011 | -0.2317 | No |
| 109 | HMGB3 | HMGB3 | 14202 | -0.012 | -0.2353 | No |
| 110 | TMEM118 | TMEM118 | 14294 | -0.012 | -0.2370 | No |
| 111 | SOX2 | SOX2 | 14311 | -0.012 | -0.2352 | No |
| 112 | ERBB3 | ERBB3 | 14471 | -0.013 | -0.2399 | No |
| 113 | YPEL1 | YPEL1 | 14618 | -0.013 | -0.2439 | No |
| 114 | DPF1 | DPF1 | 14715 | -0.014 | -0.2455 | No |
| 115 | HRASLS | HRASLS | 14793 | -0.014 | -0.2461 | No |
| 116 | MYT1 | MYT1 | 15328 | -0.016 | -0.2677 | No |
| 117 | ZNF804A | ZNF804A | 15432 | -0.017 | -0.2690 | No |
| 118 | EPHB1 | EPHB1 | 15812 | -0.018 | -0.2828 | No |
| 119 | AMOTL2 | AMOTL2 | 16000 | -0.019 | -0.2875 | No |
| 120 | REEP1 | REEP1 | 16058 | -0.019 | -0.2861 | No |
| 121 | HN1 | HN1 | 16160 | -0.020 | -0.2867 | No |
| 122 | CRMP1 | CRMP1 | 16380 | -0.021 | -0.2925 | No |
| 123 | TOX3 | TOX3 | 16422 | -0.021 | -0.2900 | No |
| 124 | PPM1E | PPM1E | 16519 | -0.021 | -0.2899 | No |
| 125 | MARCKSL1 | MARCKSL1 | 16590 | -0.022 | -0.2886 | No |
| 126 | CDK5R1 | CDK5R1 | 16855 | -0.023 | -0.2961 | No |
| 127 | NR0B1 | NR0B1 | 16982 | -0.024 | -0.2970 | No |
| 128 | CKB | CKB | 17020 | -0.024 | -0.2937 | No |
| 129 | DPYSL4 | DPYSL4 | 17265 | -0.025 | -0.2998 | No |
| 130 | SEC61A2 | SEC61A2 | 17540 | -0.026 | -0.3070 | No |
| 131 | CRB1 | CRB1 | 17541 | -0.026 | -0.3015 | No |
| 132 | C1orf106 | C1orf106 | 17766 | -0.028 | -0.3061 | No |
| 133 | CSNK1E | CSNK1E | 17942 | -0.029 | -0.3081 | No |
| 134 | RBPJ | RBPJ | 18142 | -0.030 | -0.3111 | Yes |
| 135 | MAST1 | MAST1 | 18212 | -0.030 | -0.3079 | Yes |
| 136 | DUSP26 | DUSP26 | 18351 | -0.031 | -0.3077 | Yes |
| 137 | PAFAH1B3 | PAFAH1B3 | 18371 | -0.031 | -0.3020 | Yes |
| 138 | ICK | ICK | 18543 | -0.032 | -0.3031 | Yes |
| 139 | MYST2 | MYST2 | 18726 | -0.034 | -0.3045 | Yes |
| 140 | PODXL2 | PODXL2 | 18990 | -0.035 | -0.3093 | Yes |
| 141 | WDR68 | WDR68 | 18992 | -0.035 | -0.3019 | Yes |
| 142 | ARHGEF9 | ARHGEF9 | 19238 | -0.037 | -0.3054 | Yes |
| 143 | BAI3 | BAI3 | 19366 | -0.038 | -0.3033 | Yes |
| 144 | PHF16 | PHF16 | 19395 | -0.039 | -0.2964 | Yes |
| 145 | GPR17 | GPR17 | 19501 | -0.039 | -0.2930 | Yes |
| 146 | VAX2 | VAX2 | 19506 | -0.039 | -0.2849 | Yes |
| 147 | RAB33A | RAB33A | 19565 | -0.040 | -0.2791 | Yes |
| 148 | TMEFF1 | TMEFF1 | 19736 | -0.041 | -0.2783 | Yes |
| 149 | GNG4 | GNG4 | 19883 | -0.043 | -0.2760 | Yes |
| 150 | C1QL1 | C1QL1 | 19916 | -0.044 | -0.2683 | Yes |
| 151 | BEX1 | BEX1 | 20002 | -0.045 | -0.2629 | Yes |
| 152 | NLGN3 | NLGN3 | 20159 | -0.046 | -0.2604 | Yes |
| 153 | BCAN | BCAN | 20254 | -0.048 | -0.2547 | Yes |
| 154 | PPM1D | PPM1D | 20267 | -0.048 | -0.2452 | Yes |
| 155 | MTSS1 | MTSS1 | 20289 | -0.048 | -0.2360 | Yes |
| 156 | SOX4 | SOX4 | 20350 | -0.049 | -0.2285 | Yes |
| 157 | GRIA2 | GRIA2 | 20478 | -0.051 | -0.2236 | Yes |
| 158 | NCALD | NCALD | 21072 | -0.065 | -0.2376 | Yes |
| 159 | OLIG2 | OLIG2 | 21293 | -0.075 | -0.2321 | Yes |
| 160 | FXYD6 | FXYD6 | 21404 | -0.084 | -0.2196 | Yes |
| 161 | GSTA4 | GSTA4 | 21457 | -0.091 | -0.2028 | Yes |
| 162 | SCG3 | SCG3 | 21484 | -0.096 | -0.1838 | Yes |
| 163 | DLL3 | DLL3 | 21526 | -0.105 | -0.1636 | Yes |
| 164 | CSPG5 | CSPG5 | 21531 | -0.108 | -0.1410 | Yes |
| 165 | TTYH1 | TTYH1 | 21573 | -0.137 | -0.1140 | Yes |
| 166 | GADD45G | GADD45G | 21585 | -0.158 | -0.0812 | Yes |
| 167 | C1orf61 | C1orf61 | 21592 | -0.181 | -0.0433 | Yes |
| 168 | ASCL1 | ASCL1 | 21596 | -0.206 | 0.0000 | Yes |