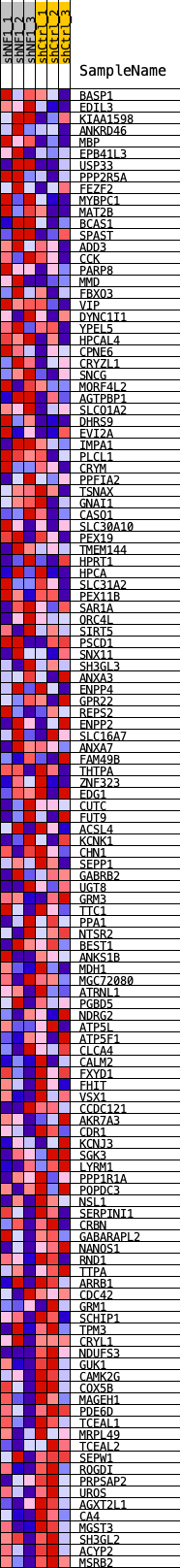
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | VERHAAK_NEU_2010 |
| Enrichment Score (ES) | -0.25262275 |
| Normalized Enrichment Score (NES) | -1.0977135 |
| Nominal p-value | 0.25906736 |
| FDR q-value | 0.38658342 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | BASP1 | BASP1 | 136 | 0.084 | 0.0198 | No |
| 2 | EDIL3 | EDIL3 | 169 | 0.080 | 0.0433 | No |
| 3 | KIAA1598 | KIAA1598 | 361 | 0.067 | 0.0553 | No |
| 4 | ANKRD46 | ANKRD46 | 412 | 0.065 | 0.0732 | No |
| 5 | MBP | MBP | 741 | 0.055 | 0.0751 | No |
| 6 | EPB41L3 | EPB41L3 | 764 | 0.055 | 0.0911 | No |
| 7 | USP33 | USP33 | 937 | 0.052 | 0.0992 | No |
| 8 | PPP2R5A | PPP2R5A | 1498 | 0.043 | 0.0865 | No |
| 9 | FEZF2 | FEZF2 | 1595 | 0.042 | 0.0952 | No |
| 10 | MYBPC1 | MYBPC1 | 1665 | 0.042 | 0.1050 | No |
| 11 | MAT2B | MAT2B | 1715 | 0.041 | 0.1155 | No |
| 12 | BCAS1 | BCAS1 | 2069 | 0.038 | 0.1108 | No |
| 13 | SPAST | SPAST | 2072 | 0.038 | 0.1225 | No |
| 14 | ADD3 | ADD3 | 2214 | 0.037 | 0.1273 | No |
| 15 | CCK | CCK | 2539 | 0.034 | 0.1228 | No |
| 16 | PARP8 | PARP8 | 2634 | 0.033 | 0.1288 | No |
| 17 | MMD | MMD | 2816 | 0.032 | 0.1304 | No |
| 18 | FBXO3 | FBXO3 | 3112 | 0.030 | 0.1260 | No |
| 19 | VIP | VIP | 3142 | 0.030 | 0.1340 | No |
| 20 | DYNC1I1 | DYNC1I1 | 3196 | 0.030 | 0.1407 | No |
| 21 | YPEL5 | YPEL5 | 3639 | 0.027 | 0.1285 | No |
| 22 | HPCAL4 | HPCAL4 | 3697 | 0.027 | 0.1341 | No |
| 23 | CPNE6 | CPNE6 | 3837 | 0.026 | 0.1357 | No |
| 24 | CRYZL1 | CRYZL1 | 3990 | 0.025 | 0.1364 | No |
| 25 | SNCG | SNCG | 4086 | 0.025 | 0.1396 | No |
| 26 | MORF4L2 | MORF4L2 | 4334 | 0.023 | 0.1353 | No |
| 27 | AGTPBP1 | AGTPBP1 | 4518 | 0.022 | 0.1338 | No |
| 28 | SLCO1A2 | SLCO1A2 | 4612 | 0.022 | 0.1363 | No |
| 29 | DHRS9 | DHRS9 | 5582 | 0.018 | 0.0966 | No |
| 30 | EVI2A | EVI2A | 5634 | 0.017 | 0.0996 | No |
| 31 | IMPA1 | IMPA1 | 5662 | 0.017 | 0.1037 | No |
| 32 | PLCL1 | PLCL1 | 5924 | 0.016 | 0.0966 | No |
| 33 | CRYM | CRYM | 6273 | 0.015 | 0.0849 | No |
| 34 | PPFIA2 | PPFIA2 | 6529 | 0.014 | 0.0773 | No |
| 35 | TSNAX | TSNAX | 6572 | 0.014 | 0.0796 | No |
| 36 | GNAI1 | GNAI1 | 6998 | 0.012 | 0.0635 | No |
| 37 | CASQ1 | CASQ1 | 7579 | 0.010 | 0.0396 | No |
| 38 | SLC30A10 | SLC30A10 | 7589 | 0.010 | 0.0422 | No |
| 39 | PEX19 | PEX19 | 7643 | 0.010 | 0.0428 | No |
| 40 | TMEM144 | TMEM144 | 7676 | 0.010 | 0.0443 | No |
| 41 | HPRT1 | HPRT1 | 7706 | 0.010 | 0.0459 | No |
| 42 | HPCA | HPCA | 7734 | 0.009 | 0.0476 | No |
| 43 | SLC31A2 | SLC31A2 | 7855 | 0.009 | 0.0448 | No |
| 44 | PEX11B | PEX11B | 8192 | 0.008 | 0.0316 | No |
| 45 | SAR1A | SAR1A | 8704 | 0.006 | 0.0098 | No |
| 46 | ORC4L | ORC4L | 8860 | 0.006 | 0.0043 | No |
| 47 | SIRT5 | SIRT5 | 8862 | 0.006 | 0.0061 | No |
| 48 | PSCD1 | PSCD1 | 9279 | 0.004 | -0.0120 | No |
| 49 | SNX11 | SNX11 | 9283 | 0.004 | -0.0108 | No |
| 50 | SH3GL3 | SH3GL3 | 9333 | 0.004 | -0.0117 | No |
| 51 | ANXA3 | ANXA3 | 9520 | 0.004 | -0.0193 | No |
| 52 | ENPP4 | ENPP4 | 9521 | 0.004 | -0.0182 | No |
| 53 | GPR22 | GPR22 | 9777 | 0.003 | -0.0292 | No |
| 54 | REPS2 | REPS2 | 9937 | 0.002 | -0.0359 | No |
| 55 | ENPP2 | ENPP2 | 9984 | 0.002 | -0.0374 | No |
| 56 | SLC16A7 | SLC16A7 | 10069 | 0.002 | -0.0408 | No |
| 57 | ANXA7 | ANXA7 | 10395 | 0.001 | -0.0557 | No |
| 58 | FAM49B | FAM49B | 10790 | -0.000 | -0.0739 | No |
| 59 | THTPA | THTPA | 10865 | -0.001 | -0.0771 | No |
| 60 | ZNF323 | ZNF323 | 11006 | -0.001 | -0.0833 | No |
| 61 | EDG1 | EDG1 | 11045 | -0.001 | -0.0847 | No |
| 62 | CUTC | CUTC | 11428 | -0.002 | -0.1017 | No |
| 63 | FUT9 | FUT9 | 11437 | -0.003 | -0.1013 | No |
| 64 | ACSL4 | ACSL4 | 11692 | -0.003 | -0.1121 | No |
| 65 | KCNK1 | KCNK1 | 11747 | -0.004 | -0.1135 | No |
| 66 | CHN1 | CHN1 | 12022 | -0.004 | -0.1249 | No |
| 67 | SEPP1 | SEPP1 | 12146 | -0.005 | -0.1291 | No |
| 68 | GABRB2 | GABRB2 | 12157 | -0.005 | -0.1280 | No |
| 69 | UGT8 | UGT8 | 12182 | -0.005 | -0.1276 | No |
| 70 | GRM3 | GRM3 | 12517 | -0.006 | -0.1413 | No |
| 71 | TTC1 | TTC1 | 12623 | -0.006 | -0.1442 | No |
| 72 | PPA1 | PPA1 | 12702 | -0.007 | -0.1457 | No |
| 73 | NTSR2 | NTSR2 | 12778 | -0.007 | -0.1471 | No |
| 74 | BEST1 | BEST1 | 12800 | -0.007 | -0.1459 | No |
| 75 | ANKS1B | ANKS1B | 12848 | -0.007 | -0.1458 | No |
| 76 | MDH1 | MDH1 | 12859 | -0.007 | -0.1441 | No |
| 77 | MGC72080 | MGC72080 | 13033 | -0.008 | -0.1497 | No |
| 78 | ATRNL1 | ATRNL1 | 13338 | -0.009 | -0.1611 | No |
| 79 | PGBD5 | PGBD5 | 13582 | -0.010 | -0.1695 | No |
| 80 | NDRG2 | NDRG2 | 13872 | -0.011 | -0.1797 | No |
| 81 | ATP5L | ATP5L | 13996 | -0.011 | -0.1820 | No |
| 82 | ATP5F1 | ATP5F1 | 14275 | -0.012 | -0.1912 | No |
| 83 | CLCA4 | CLCA4 | 14478 | -0.013 | -0.1967 | No |
| 84 | CALM2 | CALM2 | 14676 | -0.014 | -0.2016 | No |
| 85 | FXYD1 | FXYD1 | 14877 | -0.014 | -0.2065 | No |
| 86 | FHIT | FHIT | 15288 | -0.016 | -0.2207 | No |
| 87 | VSX1 | VSX1 | 15594 | -0.017 | -0.2295 | No |
| 88 | CCDC121 | CCDC121 | 16091 | -0.019 | -0.2466 | Yes |
| 89 | AKR7A3 | AKR7A3 | 16167 | -0.020 | -0.2440 | Yes |
| 90 | CDR1 | CDR1 | 16240 | -0.020 | -0.2411 | Yes |
| 91 | KCNJ3 | KCNJ3 | 16394 | -0.021 | -0.2417 | Yes |
| 92 | SGK3 | SGK3 | 16412 | -0.021 | -0.2360 | Yes |
| 93 | LYRM1 | LYRM1 | 16576 | -0.022 | -0.2369 | Yes |
| 94 | PPP1R1A | PPP1R1A | 16579 | -0.022 | -0.2302 | Yes |
| 95 | POPDC3 | POPDC3 | 16588 | -0.022 | -0.2238 | Yes |
| 96 | NSL1 | NSL1 | 16698 | -0.022 | -0.2220 | Yes |
| 97 | SERPINI1 | SERPINI1 | 16810 | -0.023 | -0.2201 | Yes |
| 98 | CRBN | CRBN | 16841 | -0.023 | -0.2144 | Yes |
| 99 | GABARAPL2 | GABARAPL2 | 16890 | -0.023 | -0.2094 | Yes |
| 100 | NANOS1 | NANOS1 | 16932 | -0.023 | -0.2041 | Yes |
| 101 | RND1 | RND1 | 17096 | -0.024 | -0.2042 | Yes |
| 102 | TTPA | TTPA | 17957 | -0.029 | -0.2353 | Yes |
| 103 | ARRB1 | ARRB1 | 18099 | -0.030 | -0.2326 | Yes |
| 104 | CDC42 | CDC42 | 18129 | -0.030 | -0.2247 | Yes |
| 105 | GRM1 | GRM1 | 18251 | -0.031 | -0.2208 | Yes |
| 106 | SCHIP1 | SCHIP1 | 18694 | -0.033 | -0.2311 | Yes |
| 107 | TPM3 | TPM3 | 18933 | -0.035 | -0.2313 | Yes |
| 108 | CRYL1 | CRYL1 | 18993 | -0.036 | -0.2230 | Yes |
| 109 | NDUFS3 | NDUFS3 | 19331 | -0.038 | -0.2269 | Yes |
| 110 | GUK1 | GUK1 | 19496 | -0.039 | -0.2223 | Yes |
| 111 | CAMK2G | CAMK2G | 20011 | -0.045 | -0.2324 | Yes |
| 112 | COX5B | COX5B | 20105 | -0.046 | -0.2226 | Yes |
| 113 | MAGEH1 | MAGEH1 | 20147 | -0.046 | -0.2101 | Yes |
| 114 | PDE6D | PDE6D | 20310 | -0.048 | -0.2027 | Yes |
| 115 | TCEAL1 | TCEAL1 | 20313 | -0.048 | -0.1877 | Yes |
| 116 | MRPL49 | MRPL49 | 20393 | -0.050 | -0.1760 | Yes |
| 117 | TCEAL2 | TCEAL2 | 20439 | -0.050 | -0.1625 | Yes |
| 118 | SEPW1 | SEPW1 | 20466 | -0.051 | -0.1479 | Yes |
| 119 | ROGDI | ROGDI | 20589 | -0.053 | -0.1372 | Yes |
| 120 | PRPSAP2 | PRPSAP2 | 20618 | -0.054 | -0.1218 | Yes |
| 121 | UROS | UROS | 20643 | -0.054 | -0.1061 | Yes |
| 122 | AGXT2L1 | AGXT2L1 | 20974 | -0.062 | -0.1023 | Yes |
| 123 | CA4 | CA4 | 21108 | -0.066 | -0.0879 | Yes |
| 124 | MGST3 | MGST3 | 21259 | -0.073 | -0.0722 | Yes |
| 125 | SH3GL2 | SH3GL2 | 21419 | -0.085 | -0.0532 | Yes |
| 126 | ACYP2 | ACYP2 | 21488 | -0.096 | -0.0267 | Yes |
| 127 | MSRB2 | MSRB2 | 21513 | -0.102 | 0.0039 | Yes |