Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | VDR_REGULON |
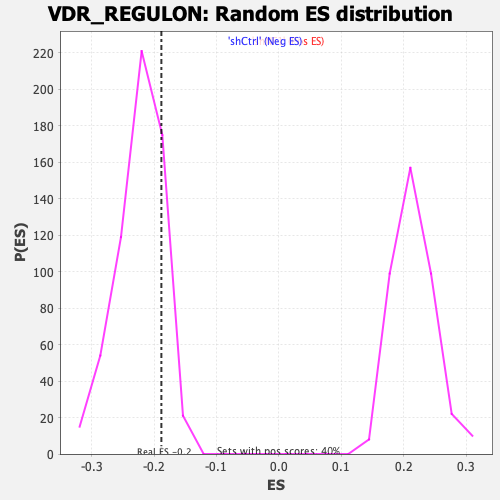
| Enrichment Score (ES) | -0.18814127 |
| Normalized Enrichment Score (NES) | -0.844639 |
| Nominal p-value | 0.8446281 |
| FDR q-value | 0.8173369 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PI15 | PI15 | 10 | 0.129 | 0.0367 | No |
| 2 | DENND3 | DENND3 | 103 | 0.088 | 0.0580 | No |
| 3 | ME1 | ME1 | 338 | 0.068 | 0.0668 | No |
| 4 | S100A8 | S100A8 | 546 | 0.060 | 0.0745 | No |
| 5 | NAV3 | NAV3 | 547 | 0.060 | 0.0918 | No |
| 6 | TNFSF9 | TNFSF9 | 638 | 0.058 | 0.1042 | No |
| 7 | ABCA7 | ABCA7 | 978 | 0.051 | 0.1031 | No |
| 8 | TCIRG1 | TCIRG1 | 1021 | 0.050 | 0.1157 | No |
| 9 | GPR39 | GPR39 | 1040 | 0.050 | 0.1292 | No |
| 10 | GCNT1 | GCNT1 | 1093 | 0.049 | 0.1410 | No |
| 11 | TFAP2C | TFAP2C | 1625 | 0.042 | 0.1284 | No |
| 12 | SERPINB7 | SERPINB7 | 1742 | 0.041 | 0.1348 | No |
| 13 | IL15RA | IL15RA | 2486 | 0.035 | 0.1102 | No |
| 14 | ZFR | ZFR | 2694 | 0.033 | 0.1101 | No |
| 15 | TNNT1 | TNNT1 | 2796 | 0.032 | 0.1147 | No |
| 16 | MPZ | MPZ | 2812 | 0.032 | 0.1233 | No |
| 17 | SMARCD2 | SMARCD2 | 2857 | 0.032 | 0.1305 | No |
| 18 | DCAF7 | DCAF7 | 3001 | 0.031 | 0.1327 | No |
| 19 | STX11 | STX11 | 3320 | 0.029 | 0.1262 | No |
| 20 | UBE2Z | UBE2Z | 3359 | 0.029 | 0.1327 | No |
| 21 | CXCL3 | CXCL3 | 3433 | 0.028 | 0.1375 | No |
| 22 | CPPED1 | CPPED1 | 3434 | 0.028 | 0.1456 | No |
| 23 | SOX11 | SOX11 | 3515 | 0.028 | 0.1499 | No |
| 24 | MSX2 | MSX2 | 3553 | 0.028 | 0.1561 | No |
| 25 | GALNT6 | GALNT6 | 3611 | 0.027 | 0.1613 | No |
| 26 | CD55 | CD55 | 3796 | 0.026 | 0.1602 | No |
| 27 | RAD50 | RAD50 | 3817 | 0.026 | 0.1668 | No |
| 28 | STEAP3 | STEAP3 | 3930 | 0.025 | 0.1689 | No |
| 29 | STARD5 | STARD5 | 4284 | 0.024 | 0.1593 | No |
| 30 | AOX1 | AOX1 | 4815 | 0.021 | 0.1406 | No |
| 31 | LAMC2 | LAMC2 | 4972 | 0.020 | 0.1392 | No |
| 32 | TMEM40 | TMEM40 | 5059 | 0.020 | 0.1410 | No |
| 33 | CASP4 | CASP4 | 5106 | 0.020 | 0.1446 | No |
| 34 | IPPK | IPPK | 5560 | 0.018 | 0.1285 | No |
| 35 | EVI2A | EVI2A | 5634 | 0.017 | 0.1301 | No |
| 36 | RAB27A | RAB27A | 6204 | 0.015 | 0.1080 | No |
| 37 | CDCP1 | CDCP1 | 6238 | 0.015 | 0.1107 | No |
| 38 | HPCAL1 | HPCAL1 | 6281 | 0.015 | 0.1130 | No |
| 39 | COL17A1 | COL17A1 | 6403 | 0.014 | 0.1114 | No |
| 40 | ABLIM3 | ABLIM3 | 6425 | 0.014 | 0.1145 | No |
| 41 | YWHAE | YWHAE | 6501 | 0.014 | 0.1150 | No |
| 42 | IQSEC2 | IQSEC2 | 6523 | 0.014 | 0.1180 | No |
| 43 | ITPR3 | ITPR3 | 6821 | 0.013 | 0.1078 | No |
| 44 | LTBR | LTBR | 6919 | 0.012 | 0.1068 | No |
| 45 | BANK1 | BANK1 | 7027 | 0.012 | 0.1053 | No |
| 46 | C3 | C3 | 7114 | 0.012 | 0.1046 | No |
| 47 | TGM2 | TGM2 | 7178 | 0.011 | 0.1049 | No |
| 48 | SH3TC1 | SH3TC1 | 7346 | 0.011 | 0.1002 | No |
| 49 | AKR1C1 | AKR1C1 | 7362 | 0.011 | 0.1026 | No |
| 50 | APOL1 | APOL1 | 7368 | 0.011 | 0.1055 | No |
| 51 | SELPLG | SELPLG | 7403 | 0.011 | 0.1069 | No |
| 52 | TBX3 | TBX3 | 7413 | 0.011 | 0.1096 | No |
| 53 | LLGL2 | LLGL2 | 7651 | 0.010 | 0.1013 | No |
| 54 | ACSL5 | ACSL5 | 7832 | 0.009 | 0.0956 | No |
| 55 | PHF7 | PHF7 | 7870 | 0.009 | 0.0964 | No |
| 56 | TNFAIP2 | TNFAIP2 | 7899 | 0.009 | 0.0977 | No |
| 57 | PLP2 | PLP2 | 8380 | 0.007 | 0.0774 | No |
| 58 | MCOLN3 | MCOLN3 | 8685 | 0.006 | 0.0651 | No |
| 59 | LRRC8E | LRRC8E | 9025 | 0.005 | 0.0508 | No |
| 60 | ABAT | ABAT | 9125 | 0.005 | 0.0475 | No |
| 61 | VNN1 | VNN1 | 9173 | 0.005 | 0.0467 | No |
| 62 | TRAPPC10 | TRAPPC10 | 9223 | 0.004 | 0.0457 | No |
| 63 | ADRB2 | ADRB2 | 9288 | 0.004 | 0.0440 | No |
| 64 | ENOX2 | ENOX2 | 9311 | 0.004 | 0.0442 | No |
| 65 | SYNGR2 | SYNGR2 | 9437 | 0.004 | 0.0395 | No |
| 66 | ABL2 | ABL2 | 9477 | 0.004 | 0.0387 | No |
| 67 | FLI1 | FLI1 | 9526 | 0.004 | 0.0375 | No |
| 68 | ALDH3A1 | ALDH3A1 | 9610 | 0.003 | 0.0346 | No |
| 69 | PVR | PVR | 9662 | 0.003 | 0.0331 | No |
| 70 | TRIM38 | TRIM38 | 9876 | 0.002 | 0.0239 | No |
| 71 | C16orf71 | C16orf71 | 9883 | 0.002 | 0.0243 | No |
| 72 | CHD7 | CHD7 | 9974 | 0.002 | 0.0207 | No |
| 73 | SRGN | SRGN | 10364 | 0.001 | 0.0028 | No |
| 74 | PRKCD | PRKCD | 10410 | 0.001 | 0.0009 | No |
| 75 | TCF12 | TCF12 | 10438 | 0.001 | -0.0002 | No |
| 76 | ASPH | ASPH | 10635 | 0.000 | -0.0093 | No |
| 77 | MID2 | MID2 | 10646 | 0.000 | -0.0098 | No |
| 78 | TNNC2 | TNNC2 | 10890 | -0.001 | -0.0209 | No |
| 79 | PRICKLE3 | PRICKLE3 | 10903 | -0.001 | -0.0212 | No |
| 80 | SP140L | SP140L | 10912 | -0.001 | -0.0213 | No |
| 81 | PKP3 | PKP3 | 10976 | -0.001 | -0.0240 | No |
| 82 | CHST15 | CHST15 | 11178 | -0.002 | -0.0329 | No |
| 83 | SDC1 | SDC1 | 11233 | -0.002 | -0.0349 | No |
| 84 | HMGA1 | HMGA1 | 11260 | -0.002 | -0.0355 | No |
| 85 | HTR7 | HTR7 | 11264 | -0.002 | -0.0351 | No |
| 86 | DNASE1L1 | DNASE1L1 | 11287 | -0.002 | -0.0356 | No |
| 87 | TRIM24 | TRIM24 | 11296 | -0.002 | -0.0353 | No |
| 88 | GMDS | GMDS | 11353 | -0.002 | -0.0373 | No |
| 89 | PTPRH | PTPRH | 11396 | -0.002 | -0.0386 | No |
| 90 | IL15 | IL15 | 11439 | -0.003 | -0.0398 | No |
| 91 | CYB5R2 | CYB5R2 | 11635 | -0.003 | -0.0480 | No |
| 92 | GPRC5A | GPRC5A | 11746 | -0.004 | -0.0521 | No |
| 93 | ADARB1 | ADARB1 | 11825 | -0.004 | -0.0546 | No |
| 94 | SPAG1 | SPAG1 | 11852 | -0.004 | -0.0547 | No |
| 95 | PIK3CD | PIK3CD | 11950 | -0.004 | -0.0580 | No |
| 96 | LTBP2 | LTBP2 | 11962 | -0.004 | -0.0573 | No |
| 97 | CDC42EP1 | CDC42EP1 | 12211 | -0.005 | -0.0674 | No |
| 98 | JPH2 | JPH2 | 12241 | -0.005 | -0.0673 | No |
| 99 | FAM114A1 | FAM114A1 | 12331 | -0.006 | -0.0698 | No |
| 100 | MME | MME | 12407 | -0.006 | -0.0717 | No |
| 101 | HLA-DQB1 | HLA-DQB1 | 12616 | -0.006 | -0.0795 | No |
| 102 | MYO1D | MYO1D | 12831 | -0.007 | -0.0874 | No |
| 103 | PTPRU | PTPRU | 13450 | -0.009 | -0.1136 | No |
| 104 | STAT6 | STAT6 | 13627 | -0.010 | -0.1190 | No |
| 105 | MAP3K1 | MAP3K1 | 13827 | -0.010 | -0.1253 | No |
| 106 | LAMB3 | LAMB3 | 13898 | -0.011 | -0.1255 | No |
| 107 | TRAF1 | TRAF1 | 13990 | -0.011 | -0.1265 | No |
| 108 | IL2RB | IL2RB | 14060 | -0.011 | -0.1265 | No |
| 109 | HMBOX1 | HMBOX1 | 14079 | -0.011 | -0.1241 | No |
| 110 | JUP | JUP | 14519 | -0.013 | -0.1408 | No |
| 111 | EXPH5 | EXPH5 | 14654 | -0.013 | -0.1432 | No |
| 112 | TNFRSF10D | TNFRSF10D | 14712 | -0.014 | -0.1419 | No |
| 113 | TUBB2A | TUBB2A | 15383 | -0.016 | -0.1684 | No |
| 114 | MAGEB2 | MAGEB2 | 15714 | -0.018 | -0.1787 | No |
| 115 | RIPK4 | RIPK4 | 15918 | -0.019 | -0.1828 | Yes |
| 116 | STEAP1 | STEAP1 | 15925 | -0.019 | -0.1777 | Yes |
| 117 | MARK1 | MARK1 | 16034 | -0.019 | -0.1772 | Yes |
| 118 | SPAG4 | SPAG4 | 16140 | -0.020 | -0.1764 | Yes |
| 119 | SPINT1 | SPINT1 | 16232 | -0.020 | -0.1748 | Yes |
| 120 | SERPINA1 | SERPINA1 | 16362 | -0.021 | -0.1749 | Yes |
| 121 | CCND3 | CCND3 | 16368 | -0.021 | -0.1691 | Yes |
| 122 | PTPRD | PTPRD | 16374 | -0.021 | -0.1634 | Yes |
| 123 | PHLPP1 | PHLPP1 | 16520 | -0.021 | -0.1640 | Yes |
| 124 | NFKBIE | NFKBIE | 16753 | -0.023 | -0.1683 | Yes |
| 125 | ID4 | ID4 | 16762 | -0.023 | -0.1621 | Yes |
| 126 | G0S2 | G0S2 | 17260 | -0.025 | -0.1781 | Yes |
| 127 | SLC9A7 | SLC9A7 | 17414 | -0.026 | -0.1778 | Yes |
| 128 | S100A2 | S100A2 | 17551 | -0.027 | -0.1765 | Yes |
| 129 | C9orf3 | C9orf3 | 17586 | -0.027 | -0.1704 | Yes |
| 130 | BCL2A1 | BCL2A1 | 17594 | -0.027 | -0.1630 | Yes |
| 131 | PDCD1LG2 | PDCD1LG2 | 17851 | -0.028 | -0.1667 | Yes |
| 132 | TRAF3IP1 | TRAF3IP1 | 17858 | -0.028 | -0.1588 | Yes |
| 133 | PLEK2 | PLEK2 | 18347 | -0.031 | -0.1725 | Yes |
| 134 | PLEKHO1 | PLEKHO1 | 18411 | -0.032 | -0.1664 | Yes |
| 135 | IL24 | IL24 | 18723 | -0.034 | -0.1711 | Yes |
| 136 | SALL2 | SALL2 | 18895 | -0.035 | -0.1691 | Yes |
| 137 | SVIL | SVIL | 19033 | -0.036 | -0.1652 | Yes |
| 138 | ICOSLG | ICOSLG | 19306 | -0.038 | -0.1669 | Yes |
| 139 | TPMT | TPMT | 19352 | -0.038 | -0.1580 | Yes |
| 140 | KYNU | KYNU | 19528 | -0.040 | -0.1547 | Yes |
| 141 | IL1R1 | IL1R1 | 19546 | -0.040 | -0.1440 | Yes |
| 142 | FMNL1 | FMNL1 | 19627 | -0.041 | -0.1361 | Yes |
| 143 | SLC10A3 | SLC10A3 | 19679 | -0.041 | -0.1266 | Yes |
| 144 | AGPAT2 | AGPAT2 | 19769 | -0.042 | -0.1187 | Yes |
| 145 | CBR1 | CBR1 | 19808 | -0.042 | -0.1083 | Yes |
| 146 | KRT7 | KRT7 | 19994 | -0.045 | -0.1040 | Yes |
| 147 | CDS1 | CDS1 | 20163 | -0.046 | -0.0985 | Yes |
| 148 | PNMA2 | PNMA2 | 20375 | -0.049 | -0.0940 | Yes |
| 149 | NCAN | NCAN | 20556 | -0.053 | -0.0873 | Yes |
| 150 | FOSL1 | FOSL1 | 20794 | -0.057 | -0.0818 | Yes |
| 151 | DOCK2 | DOCK2 | 21196 | -0.070 | -0.0803 | Yes |
| 152 | CXCL1 | CXCL1 | 21200 | -0.070 | -0.0602 | Yes |
| 153 | IGFBP6 | IGFBP6 | 21454 | -0.091 | -0.0457 | Yes |
| 154 | C1orf61 | C1orf61 | 21592 | -0.181 | 0.0002 | Yes |