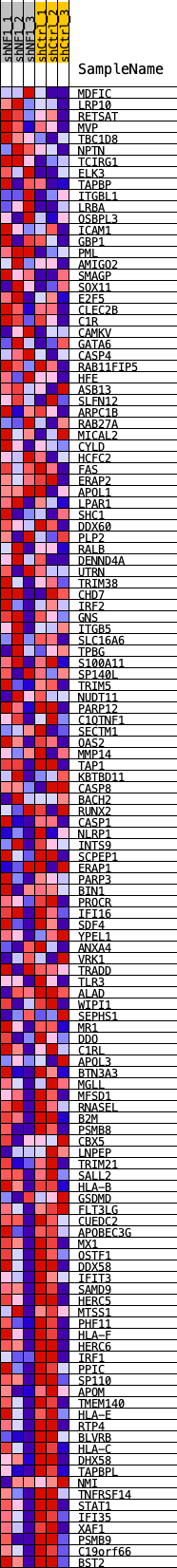
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | SP100_REGULON |
| Enrichment Score (ES) | -0.4345241 |
| Normalized Enrichment Score (NES) | -1.8772086 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.756579E-4 |
| FWER p-Value | 0.002 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MDFIC | MDFIC | 271 | 0.072 | 0.0058 | No |
| 2 | LRP10 | LRP10 | 479 | 0.062 | 0.0121 | No |
| 3 | RETSAT | RETSAT | 658 | 0.057 | 0.0184 | No |
| 4 | MVP | MVP | 711 | 0.056 | 0.0303 | No |
| 5 | TBC1D8 | TBC1D8 | 788 | 0.055 | 0.0407 | No |
| 6 | NPTN | NPTN | 878 | 0.053 | 0.0501 | No |
| 7 | TCIRG1 | TCIRG1 | 1021 | 0.050 | 0.0563 | No |
| 8 | ELK3 | ELK3 | 1072 | 0.049 | 0.0666 | No |
| 9 | TAPBP | TAPBP | 1578 | 0.043 | 0.0540 | No |
| 10 | ITGBL1 | ITGBL1 | 2047 | 0.038 | 0.0419 | No |
| 11 | LRBA | LRBA | 2261 | 0.036 | 0.0413 | No |
| 12 | OSBPL3 | OSBPL3 | 2755 | 0.033 | 0.0266 | No |
| 13 | ICAM1 | ICAM1 | 2950 | 0.031 | 0.0256 | No |
| 14 | GBP1 | GBP1 | 2993 | 0.031 | 0.0316 | No |
| 15 | PML | PML | 3150 | 0.030 | 0.0320 | No |
| 16 | AMIGO2 | AMIGO2 | 3194 | 0.030 | 0.0376 | No |
| 17 | SMAGP | SMAGP | 3414 | 0.028 | 0.0346 | No |
| 18 | SOX11 | SOX11 | 3515 | 0.028 | 0.0370 | No |
| 19 | E2F5 | E2F5 | 3627 | 0.027 | 0.0387 | No |
| 20 | CLEC2B | CLEC2B | 4145 | 0.024 | 0.0208 | No |
| 21 | C1R | C1R | 4214 | 0.024 | 0.0238 | No |
| 22 | CAMKV | CAMKV | 4489 | 0.023 | 0.0168 | No |
| 23 | GATA6 | GATA6 | 4962 | 0.020 | 0.0000 | No |
| 24 | CASP4 | CASP4 | 5106 | 0.020 | -0.0016 | No |
| 25 | RAB11FIP5 | RAB11FIP5 | 5143 | 0.020 | 0.0017 | No |
| 26 | HFE | HFE | 5423 | 0.018 | -0.0066 | No |
| 27 | ASB13 | ASB13 | 5884 | 0.016 | -0.0238 | No |
| 28 | SLFN12 | SLFN12 | 5907 | 0.016 | -0.0207 | No |
| 29 | ARPC1B | ARPC1B | 6192 | 0.015 | -0.0301 | No |
| 30 | RAB27A | RAB27A | 6204 | 0.015 | -0.0267 | No |
| 31 | MICAL2 | MICAL2 | 6513 | 0.014 | -0.0376 | No |
| 32 | CYLD | CYLD | 6516 | 0.014 | -0.0341 | No |
| 33 | HCFC2 | HCFC2 | 6633 | 0.013 | -0.0361 | No |
| 34 | FAS | FAS | 6689 | 0.013 | -0.0353 | No |
| 35 | ERAP2 | ERAP2 | 6928 | 0.012 | -0.0433 | No |
| 36 | APOL1 | APOL1 | 7368 | 0.011 | -0.0610 | No |
| 37 | LPAR1 | LPAR1 | 7876 | 0.009 | -0.0824 | No |
| 38 | SHC1 | SHC1 | 8045 | 0.008 | -0.0880 | No |
| 39 | DDX60 | DDX60 | 8367 | 0.007 | -0.1011 | No |
| 40 | PLP2 | PLP2 | 8380 | 0.007 | -0.0998 | No |
| 41 | RALB | RALB | 8436 | 0.007 | -0.1005 | No |
| 42 | DENND4A | DENND4A | 8647 | 0.006 | -0.1087 | No |
| 43 | UTRN | UTRN | 9466 | 0.004 | -0.1458 | No |
| 44 | TRIM38 | TRIM38 | 9876 | 0.002 | -0.1643 | No |
| 45 | CHD7 | CHD7 | 9974 | 0.002 | -0.1683 | No |
| 46 | IRF2 | IRF2 | 9976 | 0.002 | -0.1678 | No |
| 47 | GNS | GNS | 10291 | 0.001 | -0.1821 | No |
| 48 | ITGB5 | ITGB5 | 10457 | 0.001 | -0.1897 | No |
| 49 | SLC16A6 | SLC16A6 | 10468 | 0.001 | -0.1900 | No |
| 50 | TPBG | TPBG | 10604 | 0.000 | -0.1962 | No |
| 51 | S100A11 | S100A11 | 10703 | -0.000 | -0.2007 | No |
| 52 | SP140L | SP140L | 10912 | -0.001 | -0.2102 | No |
| 53 | TRIM5 | TRIM5 | 11347 | -0.002 | -0.2299 | No |
| 54 | NUDT11 | NUDT11 | 11624 | -0.003 | -0.2419 | No |
| 55 | PARP12 | PARP12 | 11918 | -0.004 | -0.2545 | No |
| 56 | C1QTNF1 | C1QTNF1 | 11992 | -0.004 | -0.2568 | No |
| 57 | SECTM1 | SECTM1 | 12070 | -0.005 | -0.2592 | No |
| 58 | OAS2 | OAS2 | 12174 | -0.005 | -0.2628 | No |
| 59 | MMP14 | MMP14 | 12245 | -0.005 | -0.2647 | No |
| 60 | TAP1 | TAP1 | 12289 | -0.005 | -0.2653 | No |
| 61 | KBTBD11 | KBTBD11 | 12489 | -0.006 | -0.2731 | No |
| 62 | CASP8 | CASP8 | 12893 | -0.007 | -0.2900 | No |
| 63 | BACH2 | BACH2 | 12964 | -0.008 | -0.2913 | No |
| 64 | RUNX2 | RUNX2 | 13175 | -0.008 | -0.2990 | No |
| 65 | CASP1 | CASP1 | 13302 | -0.009 | -0.3026 | No |
| 66 | NLRP1 | NLRP1 | 13867 | -0.011 | -0.3262 | No |
| 67 | INTS9 | INTS9 | 13879 | -0.011 | -0.3240 | No |
| 68 | SCPEP1 | SCPEP1 | 13899 | -0.011 | -0.3222 | No |
| 69 | ERAP1 | ERAP1 | 13923 | -0.011 | -0.3205 | No |
| 70 | PARP3 | PARP3 | 13963 | -0.011 | -0.3195 | No |
| 71 | BIN1 | BIN1 | 13979 | -0.011 | -0.3174 | No |
| 72 | PROCR | PROCR | 14056 | -0.011 | -0.3181 | No |
| 73 | IFI16 | IFI16 | 14371 | -0.012 | -0.3296 | No |
| 74 | SDF4 | SDF4 | 14481 | -0.013 | -0.3314 | No |
| 75 | YPEL1 | YPEL1 | 14618 | -0.013 | -0.3343 | No |
| 76 | ANXA4 | ANXA4 | 14631 | -0.013 | -0.3314 | No |
| 77 | VRK1 | VRK1 | 14755 | -0.014 | -0.3336 | No |
| 78 | TRADD | TRADD | 14833 | -0.014 | -0.3336 | No |
| 79 | TLR3 | TLR3 | 15001 | -0.015 | -0.3376 | No |
| 80 | ALAD | ALAD | 15243 | -0.016 | -0.3448 | No |
| 81 | WIPI1 | WIPI1 | 15356 | -0.016 | -0.3459 | No |
| 82 | SEPHS1 | SEPHS1 | 15849 | -0.018 | -0.3641 | No |
| 83 | MR1 | MR1 | 15904 | -0.019 | -0.3619 | No |
| 84 | DDO | DDO | 16097 | -0.019 | -0.3659 | No |
| 85 | C1RL | C1RL | 16390 | -0.021 | -0.3741 | No |
| 86 | APOL3 | APOL3 | 16408 | -0.021 | -0.3696 | No |
| 87 | BTN3A3 | BTN3A3 | 16601 | -0.022 | -0.3730 | No |
| 88 | MGLL | MGLL | 16941 | -0.023 | -0.3828 | No |
| 89 | MFSD1 | MFSD1 | 16964 | -0.023 | -0.3778 | No |
| 90 | RNASEL | RNASEL | 17483 | -0.026 | -0.3952 | No |
| 91 | B2M | B2M | 17865 | -0.028 | -0.4057 | No |
| 92 | PSMB8 | PSMB8 | 18363 | -0.031 | -0.4209 | No |
| 93 | CBX5 | CBX5 | 18368 | -0.031 | -0.4130 | No |
| 94 | LNPEP | LNPEP | 18551 | -0.032 | -0.4132 | No |
| 95 | TRIM21 | TRIM21 | 18682 | -0.033 | -0.4108 | No |
| 96 | SALL2 | SALL2 | 18895 | -0.035 | -0.4118 | No |
| 97 | HLA-B | HLA-B | 18926 | -0.035 | -0.4042 | No |
| 98 | GSDMD | GSDMD | 19445 | -0.039 | -0.4184 | No |
| 99 | FLT3LG | FLT3LG | 19792 | -0.042 | -0.4238 | Yes |
| 100 | CUEDC2 | CUEDC2 | 19824 | -0.042 | -0.4144 | Yes |
| 101 | APOBEC3G | APOBEC3G | 19920 | -0.044 | -0.4076 | Yes |
| 102 | MX1 | MX1 | 19984 | -0.044 | -0.3992 | Yes |
| 103 | OSTF1 | OSTF1 | 20018 | -0.045 | -0.3893 | Yes |
| 104 | DDX58 | DDX58 | 20077 | -0.045 | -0.3804 | Yes |
| 105 | IFIT3 | IFIT3 | 20082 | -0.045 | -0.3690 | Yes |
| 106 | SAMD9 | SAMD9 | 20134 | -0.046 | -0.3596 | Yes |
| 107 | HERC5 | HERC5 | 20185 | -0.047 | -0.3501 | Yes |
| 108 | MTSS1 | MTSS1 | 20289 | -0.048 | -0.3426 | Yes |
| 109 | PHF11 | PHF11 | 20428 | -0.050 | -0.3362 | Yes |
| 110 | HLA-F | HLA-F | 20653 | -0.054 | -0.3327 | Yes |
| 111 | HERC6 | HERC6 | 20663 | -0.055 | -0.3192 | Yes |
| 112 | IRF1 | IRF1 | 20690 | -0.055 | -0.3063 | Yes |
| 113 | PPIC | PPIC | 20717 | -0.056 | -0.2933 | Yes |
| 114 | SP110 | SP110 | 20846 | -0.058 | -0.2844 | Yes |
| 115 | APOM | APOM | 20908 | -0.060 | -0.2719 | Yes |
| 116 | TMEM140 | TMEM140 | 20922 | -0.060 | -0.2571 | Yes |
| 117 | HLA-E | HLA-E | 20930 | -0.061 | -0.2419 | Yes |
| 118 | RTP4 | RTP4 | 21020 | -0.063 | -0.2299 | Yes |
| 119 | BLVRB | BLVRB | 21104 | -0.066 | -0.2168 | Yes |
| 120 | HLA-C | HLA-C | 21127 | -0.067 | -0.2007 | Yes |
| 121 | DHX58 | DHX58 | 21153 | -0.068 | -0.1844 | Yes |
| 122 | TAPBPL | TAPBPL | 21272 | -0.074 | -0.1710 | Yes |
| 123 | NMI | NMI | 21290 | -0.075 | -0.1527 | Yes |
| 124 | TNFRSF14 | TNFRSF14 | 21312 | -0.076 | -0.1343 | Yes |
| 125 | STAT1 | STAT1 | 21320 | -0.076 | -0.1152 | Yes |
| 126 | IFI35 | IFI35 | 21374 | -0.081 | -0.0970 | Yes |
| 127 | XAF1 | XAF1 | 21402 | -0.084 | -0.0769 | Yes |
| 128 | PSMB9 | PSMB9 | 21406 | -0.084 | -0.0555 | Yes |
| 129 | C19orf66 | C19orf66 | 21434 | -0.087 | -0.0345 | Yes |
| 130 | BST2 | BST2 | 21587 | -0.164 | 0.0004 | Yes |