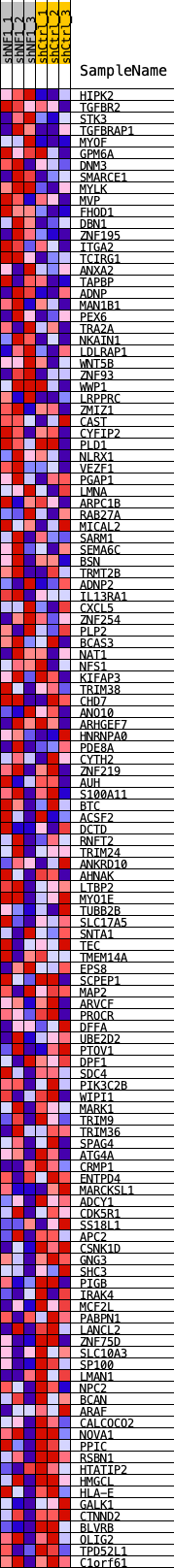
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | SOX11_REGULON |
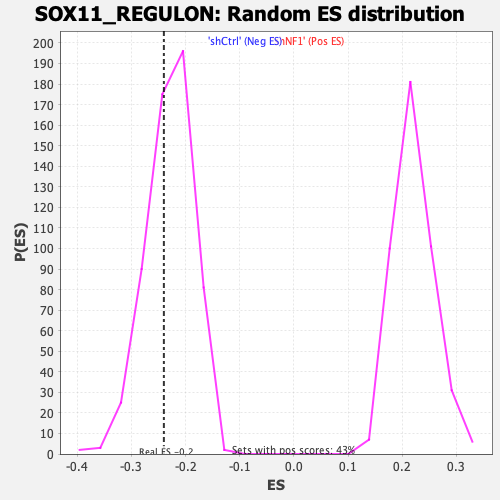
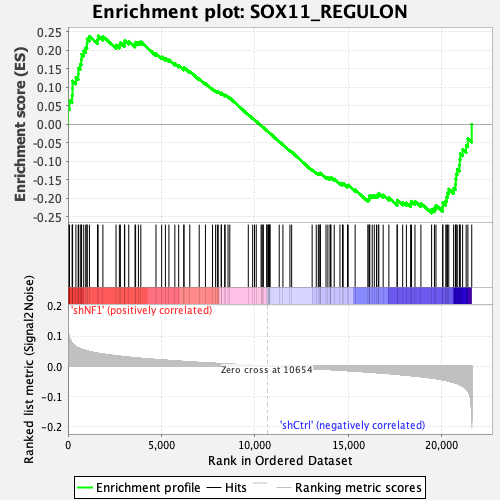
| Enrichment Score (ES) | -0.23992704 |
| Normalized Enrichment Score (NES) | -1.0461549 |
| Nominal p-value | 0.35714287 |
| FDR q-value | 0.43592647 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | HIPK2 | HIPK2 | 2 | 0.164 | 0.0437 | Yes |
| 2 | TGFBR2 | TGFBR2 | 80 | 0.093 | 0.0649 | Yes |
| 3 | STK3 | STK3 | 220 | 0.076 | 0.0787 | Yes |
| 4 | TGFBRAP1 | TGFBRAP1 | 241 | 0.075 | 0.0977 | Yes |
| 5 | MYOF | MYOF | 245 | 0.074 | 0.1174 | Yes |
| 6 | GPM6A | GPM6A | 422 | 0.065 | 0.1266 | Yes |
| 7 | DNM3 | DNM3 | 548 | 0.060 | 0.1368 | Yes |
| 8 | SMARCE1 | SMARCE1 | 556 | 0.060 | 0.1524 | Yes |
| 9 | MYLK | MYLK | 654 | 0.057 | 0.1632 | Yes |
| 10 | MVP | MVP | 711 | 0.056 | 0.1756 | Yes |
| 11 | FHOD1 | FHOD1 | 726 | 0.056 | 0.1898 | Yes |
| 12 | DBN1 | DBN1 | 840 | 0.054 | 0.1989 | Yes |
| 13 | ZNF195 | ZNF195 | 948 | 0.052 | 0.2077 | Yes |
| 14 | ITGA2 | ITGA2 | 1015 | 0.050 | 0.2180 | Yes |
| 15 | TCIRG1 | TCIRG1 | 1021 | 0.050 | 0.2312 | Yes |
| 16 | ANXA2 | ANXA2 | 1146 | 0.048 | 0.2383 | Yes |
| 17 | TAPBP | TAPBP | 1578 | 0.043 | 0.2296 | Yes |
| 18 | ADNP | ADNP | 1610 | 0.042 | 0.2395 | Yes |
| 19 | MAN1B1 | MAN1B1 | 1866 | 0.040 | 0.2382 | No |
| 20 | PEX6 | PEX6 | 2568 | 0.034 | 0.2146 | No |
| 21 | TRA2A | TRA2A | 2748 | 0.033 | 0.2150 | No |
| 22 | NKAIN1 | NKAIN1 | 2801 | 0.032 | 0.2212 | No |
| 23 | LDLRAP1 | LDLRAP1 | 3019 | 0.031 | 0.2194 | No |
| 24 | WNT5B | WNT5B | 3033 | 0.031 | 0.2270 | No |
| 25 | ZNF93 | ZNF93 | 3249 | 0.029 | 0.2248 | No |
| 26 | WWP1 | WWP1 | 3584 | 0.027 | 0.2166 | No |
| 27 | LRPPRC | LRPPRC | 3615 | 0.027 | 0.2224 | No |
| 28 | ZMIZ1 | ZMIZ1 | 3766 | 0.026 | 0.2224 | No |
| 29 | CAST | CAST | 3894 | 0.026 | 0.2234 | No |
| 30 | CYFIP2 | CYFIP2 | 4705 | 0.022 | 0.1914 | No |
| 31 | PLD1 | PLD1 | 5008 | 0.020 | 0.1827 | No |
| 32 | NLRX1 | NLRX1 | 5209 | 0.019 | 0.1786 | No |
| 33 | VEZF1 | VEZF1 | 5397 | 0.018 | 0.1748 | No |
| 34 | PGAP1 | PGAP1 | 5714 | 0.017 | 0.1646 | No |
| 35 | LMNA | LMNA | 5919 | 0.016 | 0.1595 | No |
| 36 | ARPC1B | ARPC1B | 6192 | 0.015 | 0.1508 | No |
| 37 | RAB27A | RAB27A | 6204 | 0.015 | 0.1543 | No |
| 38 | MICAL2 | MICAL2 | 6513 | 0.014 | 0.1437 | No |
| 39 | SARM1 | SARM1 | 7016 | 0.012 | 0.1234 | No |
| 40 | SEMA6C | SEMA6C | 7350 | 0.011 | 0.1108 | No |
| 41 | BSN | BSN | 7729 | 0.009 | 0.0957 | No |
| 42 | TRMT2B | TRMT2B | 7894 | 0.009 | 0.0905 | No |
| 43 | ADNP2 | ADNP2 | 7989 | 0.009 | 0.0884 | No |
| 44 | IL13RA1 | IL13RA1 | 8033 | 0.008 | 0.0886 | No |
| 45 | CXCL5 | CXCL5 | 8194 | 0.008 | 0.0833 | No |
| 46 | ZNF254 | ZNF254 | 8202 | 0.008 | 0.0850 | No |
| 47 | PLP2 | PLP2 | 8380 | 0.007 | 0.0788 | No |
| 48 | BCAS3 | BCAS3 | 8401 | 0.007 | 0.0798 | No |
| 49 | NAT1 | NAT1 | 8558 | 0.007 | 0.0743 | No |
| 50 | NFS1 | NFS1 | 8652 | 0.006 | 0.0717 | No |
| 51 | KIFAP3 | KIFAP3 | 9642 | 0.003 | 0.0264 | No |
| 52 | TRIM38 | TRIM38 | 9876 | 0.002 | 0.0162 | No |
| 53 | CHD7 | CHD7 | 9974 | 0.002 | 0.0123 | No |
| 54 | ANO10 | ANO10 | 10068 | 0.002 | 0.0084 | No |
| 55 | ARHGEF7 | ARHGEF7 | 10327 | 0.001 | -0.0033 | No |
| 56 | HNRNPA0 | HNRNPA0 | 10373 | 0.001 | -0.0052 | No |
| 57 | PDE8A | PDE8A | 10425 | 0.001 | -0.0074 | No |
| 58 | CYTH2 | CYTH2 | 10444 | 0.001 | -0.0080 | No |
| 59 | ZNF219 | ZNF219 | 10626 | 0.000 | -0.0165 | No |
| 60 | AUH | AUH | 10647 | 0.000 | -0.0174 | No |
| 61 | S100A11 | S100A11 | 10703 | -0.000 | -0.0199 | No |
| 62 | BTC | BTC | 10725 | -0.000 | -0.0208 | No |
| 63 | ACSF2 | ACSF2 | 10772 | -0.000 | -0.0229 | No |
| 64 | DCTD | DCTD | 10794 | -0.000 | -0.0237 | No |
| 65 | RNFT2 | RNFT2 | 10803 | -0.000 | -0.0240 | No |
| 66 | TRIM24 | TRIM24 | 11296 | -0.002 | -0.0463 | No |
| 67 | ANKRD10 | ANKRD10 | 11494 | -0.003 | -0.0548 | No |
| 68 | AHNAK | AHNAK | 11869 | -0.004 | -0.0712 | No |
| 69 | LTBP2 | LTBP2 | 11962 | -0.004 | -0.0743 | No |
| 70 | MYO1E | MYO1E | 13057 | -0.008 | -0.1232 | No |
| 71 | TUBB2B | TUBB2B | 13277 | -0.009 | -0.1311 | No |
| 72 | SLC17A5 | SLC17A5 | 13387 | -0.009 | -0.1338 | No |
| 73 | SNTA1 | SNTA1 | 13429 | -0.009 | -0.1333 | No |
| 74 | TEC | TEC | 13487 | -0.009 | -0.1334 | No |
| 75 | TMEM14A | TMEM14A | 13498 | -0.009 | -0.1314 | No |
| 76 | EPS8 | EPS8 | 13798 | -0.010 | -0.1426 | No |
| 77 | SCPEP1 | SCPEP1 | 13899 | -0.011 | -0.1444 | No |
| 78 | MAP2 | MAP2 | 14012 | -0.011 | -0.1467 | No |
| 79 | ARVCF | ARVCF | 14051 | -0.011 | -0.1454 | No |
| 80 | PROCR | PROCR | 14056 | -0.011 | -0.1426 | No |
| 81 | DFFA | DFFA | 14236 | -0.012 | -0.1478 | No |
| 82 | UBE2D2 | UBE2D2 | 14545 | -0.013 | -0.1586 | No |
| 83 | PTOV1 | PTOV1 | 14681 | -0.014 | -0.1613 | No |
| 84 | DPF1 | DPF1 | 14715 | -0.014 | -0.1592 | No |
| 85 | SDC4 | SDC4 | 14948 | -0.015 | -0.1661 | No |
| 86 | PIK3C2B | PIK3C2B | 14984 | -0.015 | -0.1638 | No |
| 87 | WIPI1 | WIPI1 | 15356 | -0.016 | -0.1767 | No |
| 88 | MARK1 | MARK1 | 16034 | -0.019 | -0.2031 | No |
| 89 | TRIM9 | TRIM9 | 16098 | -0.019 | -0.2009 | No |
| 90 | TRIM36 | TRIM36 | 16112 | -0.020 | -0.1963 | No |
| 91 | SPAG4 | SPAG4 | 16140 | -0.020 | -0.1923 | No |
| 92 | ATG4A | ATG4A | 16269 | -0.020 | -0.1928 | No |
| 93 | CRMP1 | CRMP1 | 16380 | -0.021 | -0.1924 | No |
| 94 | ENTPD4 | ENTPD4 | 16498 | -0.021 | -0.1921 | No |
| 95 | MARCKSL1 | MARCKSL1 | 16590 | -0.022 | -0.1906 | No |
| 96 | ADCY1 | ADCY1 | 16623 | -0.022 | -0.1862 | No |
| 97 | CDK5R1 | CDK5R1 | 16855 | -0.023 | -0.1908 | No |
| 98 | SS18L1 | SS18L1 | 17161 | -0.024 | -0.1985 | No |
| 99 | APC2 | APC2 | 17604 | -0.027 | -0.2119 | No |
| 100 | CSNK1D | CSNK1D | 17606 | -0.027 | -0.2048 | No |
| 101 | GNG3 | GNG3 | 17899 | -0.029 | -0.2107 | No |
| 102 | SHC3 | SHC3 | 18098 | -0.030 | -0.2120 | No |
| 103 | PIGB | PIGB | 18323 | -0.031 | -0.2141 | No |
| 104 | IRAK4 | IRAK4 | 18373 | -0.031 | -0.2080 | No |
| 105 | MCF2L | MCF2L | 18556 | -0.032 | -0.2078 | No |
| 106 | PABPN1 | PABPN1 | 18874 | -0.035 | -0.2133 | No |
| 107 | LANCL2 | LANCL2 | 19446 | -0.039 | -0.2295 | No |
| 108 | ZNF75D | ZNF75D | 19598 | -0.040 | -0.2258 | No |
| 109 | SLC10A3 | SLC10A3 | 19679 | -0.041 | -0.2186 | No |
| 110 | SP100 | SP100 | 20037 | -0.045 | -0.2232 | No |
| 111 | LMAN1 | LMAN1 | 20047 | -0.045 | -0.2116 | No |
| 112 | NPC2 | NPC2 | 20205 | -0.047 | -0.2064 | No |
| 113 | BCAN | BCAN | 20254 | -0.048 | -0.1959 | No |
| 114 | ARAF | ARAF | 20300 | -0.048 | -0.1851 | No |
| 115 | CALCOCO2 | CALCOCO2 | 20364 | -0.049 | -0.1749 | No |
| 116 | NOVA1 | NOVA1 | 20622 | -0.054 | -0.1725 | No |
| 117 | PPIC | PPIC | 20717 | -0.056 | -0.1620 | No |
| 118 | RSBN1 | RSBN1 | 20735 | -0.056 | -0.1479 | No |
| 119 | HTATIP2 | HTATIP2 | 20761 | -0.057 | -0.1339 | No |
| 120 | HMGCL | HMGCL | 20817 | -0.058 | -0.1211 | No |
| 121 | HLA-E | HLA-E | 20930 | -0.061 | -0.1101 | No |
| 122 | GALK1 | GALK1 | 20950 | -0.061 | -0.0946 | No |
| 123 | CTNND2 | CTNND2 | 20977 | -0.062 | -0.0793 | No |
| 124 | BLVRB | BLVRB | 21104 | -0.066 | -0.0674 | No |
| 125 | OLIG2 | OLIG2 | 21293 | -0.075 | -0.0562 | No |
| 126 | TPD52L1 | TPD52L1 | 21385 | -0.082 | -0.0386 | No |
| 127 | C1orf61 | C1orf61 | 21592 | -0.181 | 0.0002 | No |