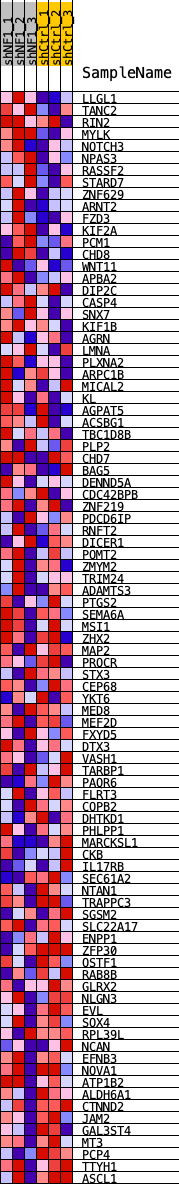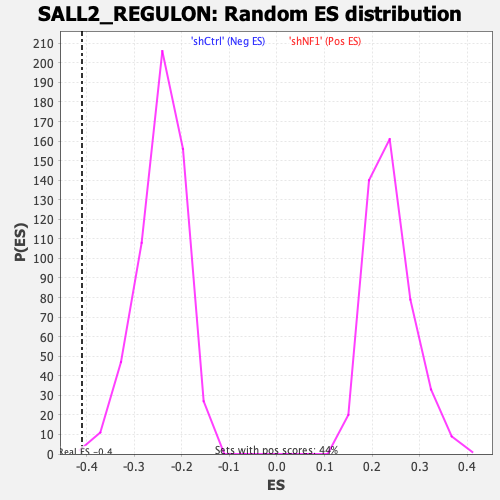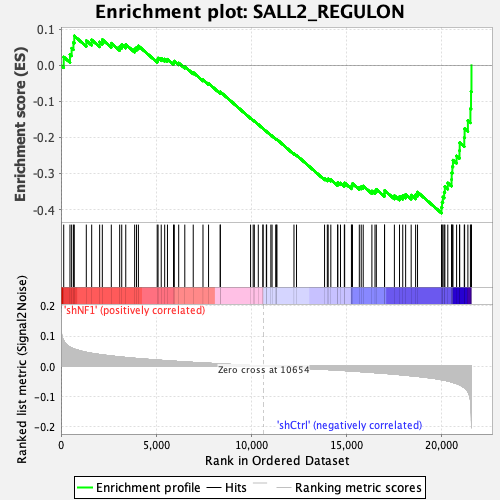
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | SALL2_REGULON |
| Enrichment Score (ES) | -0.4101428 |
| Normalized Enrichment Score (NES) | -1.6800829 |
| Nominal p-value | 0.0017953322 |
| FDR q-value | 0.005655233 |
| FWER p-Value | 0.041 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LLGL1 | LLGL1 | 143 | 0.084 | 0.0232 | No |
| 2 | TANC2 | TANC2 | 472 | 0.062 | 0.0302 | No |
| 3 | RIN2 | RIN2 | 560 | 0.060 | 0.0475 | No |
| 4 | MYLK | MYLK | 654 | 0.057 | 0.0637 | No |
| 5 | NOTCH3 | NOTCH3 | 698 | 0.056 | 0.0818 | No |
| 6 | NPAS3 | NPAS3 | 1331 | 0.046 | 0.0687 | No |
| 7 | RASSF2 | RASSF2 | 1614 | 0.042 | 0.0706 | No |
| 8 | STARD7 | STARD7 | 2033 | 0.038 | 0.0649 | No |
| 9 | ZNF629 | ZNF629 | 2174 | 0.037 | 0.0716 | No |
| 10 | ARNT2 | ARNT2 | 2647 | 0.033 | 0.0616 | No |
| 11 | FZD3 | FZD3 | 3087 | 0.030 | 0.0520 | No |
| 12 | KIF2A | KIF2A | 3195 | 0.030 | 0.0577 | No |
| 13 | PCM1 | PCM1 | 3407 | 0.028 | 0.0579 | No |
| 14 | CHD8 | CHD8 | 3875 | 0.026 | 0.0454 | No |
| 15 | WNT11 | WNT11 | 3973 | 0.025 | 0.0499 | No |
| 16 | APBA2 | APBA2 | 4072 | 0.025 | 0.0541 | No |
| 17 | DIP2C | DIP2C | 5061 | 0.020 | 0.0153 | No |
| 18 | CASP4 | CASP4 | 5106 | 0.020 | 0.0203 | No |
| 19 | SNX7 | SNX7 | 5273 | 0.019 | 0.0194 | No |
| 20 | KIF1B | KIF1B | 5455 | 0.018 | 0.0175 | No |
| 21 | AGRN | AGRN | 5601 | 0.017 | 0.0170 | No |
| 22 | LMNA | LMNA | 5919 | 0.016 | 0.0080 | No |
| 23 | PLXNA2 | PLXNA2 | 5961 | 0.016 | 0.0119 | No |
| 24 | ARPC1B | ARPC1B | 6192 | 0.015 | 0.0066 | No |
| 25 | MICAL2 | MICAL2 | 6513 | 0.014 | -0.0034 | No |
| 26 | KL | KL | 6957 | 0.012 | -0.0197 | No |
| 27 | AGPAT5 | AGPAT5 | 7470 | 0.010 | -0.0398 | No |
| 28 | ACSBG1 | ACSBG1 | 7763 | 0.009 | -0.0500 | No |
| 29 | TBC1D8B | TBC1D8B | 8379 | 0.007 | -0.0760 | No |
| 30 | PLP2 | PLP2 | 8380 | 0.007 | -0.0734 | No |
| 31 | CHD7 | CHD7 | 9974 | 0.002 | -0.1467 | No |
| 32 | BAG5 | BAG5 | 10111 | 0.002 | -0.1524 | No |
| 33 | DENND5A | DENND5A | 10171 | 0.001 | -0.1546 | No |
| 34 | CDC42BPB | CDC42BPB | 10380 | 0.001 | -0.1640 | No |
| 35 | ZNF219 | ZNF219 | 10626 | 0.000 | -0.1754 | No |
| 36 | PDCD6IP | PDCD6IP | 10627 | 0.000 | -0.1753 | No |
| 37 | RNFT2 | RNFT2 | 10803 | -0.000 | -0.1833 | No |
| 38 | DICER1 | DICER1 | 10819 | -0.001 | -0.1838 | No |
| 39 | POMT2 | POMT2 | 11041 | -0.001 | -0.1936 | No |
| 40 | ZMYM2 | ZMYM2 | 11104 | -0.001 | -0.1960 | No |
| 41 | TRIM24 | TRIM24 | 11296 | -0.002 | -0.2042 | No |
| 42 | ADAMTS3 | ADAMTS3 | 11346 | -0.002 | -0.2057 | No |
| 43 | PTGS2 | PTGS2 | 11356 | -0.002 | -0.2053 | No |
| 44 | SEMA6A | SEMA6A | 12257 | -0.005 | -0.2453 | No |
| 45 | MSI1 | MSI1 | 12391 | -0.006 | -0.2494 | No |
| 46 | ZHX2 | ZHX2 | 13866 | -0.011 | -0.3142 | No |
| 47 | MAP2 | MAP2 | 14012 | -0.011 | -0.3170 | No |
| 48 | PROCR | PROCR | 14056 | -0.011 | -0.3150 | No |
| 49 | STX3 | STX3 | 14196 | -0.012 | -0.3172 | No |
| 50 | CEP68 | CEP68 | 14552 | -0.013 | -0.3291 | No |
| 51 | YKT6 | YKT6 | 14564 | -0.013 | -0.3249 | No |
| 52 | MED8 | MED8 | 14705 | -0.014 | -0.3265 | No |
| 53 | MEF2D | MEF2D | 14907 | -0.014 | -0.3307 | No |
| 54 | FXYD5 | FXYD5 | 14924 | -0.014 | -0.3263 | No |
| 55 | DTX3 | DTX3 | 15280 | -0.016 | -0.3371 | No |
| 56 | VASH1 | VASH1 | 15305 | -0.016 | -0.3325 | No |
| 57 | TARBP1 | TARBP1 | 15334 | -0.016 | -0.3280 | No |
| 58 | PAQR6 | PAQR6 | 15695 | -0.018 | -0.3385 | No |
| 59 | FLRT3 | FLRT3 | 15795 | -0.018 | -0.3366 | No |
| 60 | COPB2 | COPB2 | 15900 | -0.019 | -0.3348 | No |
| 61 | DHTKD1 | DHTKD1 | 16357 | -0.021 | -0.3486 | No |
| 62 | PHLPP1 | PHLPP1 | 16520 | -0.021 | -0.3485 | No |
| 63 | MARCKSL1 | MARCKSL1 | 16590 | -0.022 | -0.3439 | No |
| 64 | CKB | CKB | 17020 | -0.024 | -0.3554 | No |
| 65 | IL17RB | IL17RB | 17025 | -0.024 | -0.3471 | No |
| 66 | SEC61A2 | SEC61A2 | 17540 | -0.026 | -0.3616 | No |
| 67 | NTAN1 | NTAN1 | 17810 | -0.028 | -0.3641 | No |
| 68 | TRAPPC3 | TRAPPC3 | 17980 | -0.029 | -0.3615 | No |
| 69 | SGSM2 | SGSM2 | 18132 | -0.030 | -0.3579 | No |
| 70 | SLC22A17 | SLC22A17 | 18423 | -0.032 | -0.3600 | No |
| 71 | ENPP1 | ENPP1 | 18657 | -0.033 | -0.3590 | No |
| 72 | ZFP30 | ZFP30 | 18758 | -0.034 | -0.3516 | No |
| 73 | OSTF1 | OSTF1 | 20018 | -0.045 | -0.3941 | Yes |
| 74 | RAB8B | RAB8B | 20048 | -0.045 | -0.3794 | Yes |
| 75 | GLRX2 | GLRX2 | 20093 | -0.046 | -0.3652 | Yes |
| 76 | NLGN3 | NLGN3 | 20159 | -0.046 | -0.3517 | Yes |
| 77 | EVL | EVL | 20196 | -0.047 | -0.3367 | Yes |
| 78 | SOX4 | SOX4 | 20350 | -0.049 | -0.3263 | Yes |
| 79 | RPL39L | RPL39L | 20547 | -0.052 | -0.3167 | Yes |
| 80 | NCAN | NCAN | 20556 | -0.053 | -0.2983 | Yes |
| 81 | EFNB3 | EFNB3 | 20590 | -0.053 | -0.2809 | Yes |
| 82 | NOVA1 | NOVA1 | 20622 | -0.054 | -0.2631 | Yes |
| 83 | ATP1B2 | ATP1B2 | 20813 | -0.058 | -0.2514 | Yes |
| 84 | ALDH6A1 | ALDH6A1 | 20966 | -0.062 | -0.2364 | Yes |
| 85 | CTNND2 | CTNND2 | 20977 | -0.062 | -0.2147 | Yes |
| 86 | JAM2 | JAM2 | 21217 | -0.071 | -0.2005 | Yes |
| 87 | GAL3ST4 | GAL3ST4 | 21235 | -0.072 | -0.1756 | Yes |
| 88 | MT3 | MT3 | 21407 | -0.084 | -0.1535 | Yes |
| 89 | PCP4 | PCP4 | 21540 | -0.111 | -0.1200 | Yes |
| 90 | TTYH1 | TTYH1 | 21573 | -0.137 | -0.0725 | Yes |
| 91 | ASCL1 | ASCL1 | 21596 | -0.206 | 0.0000 | Yes |