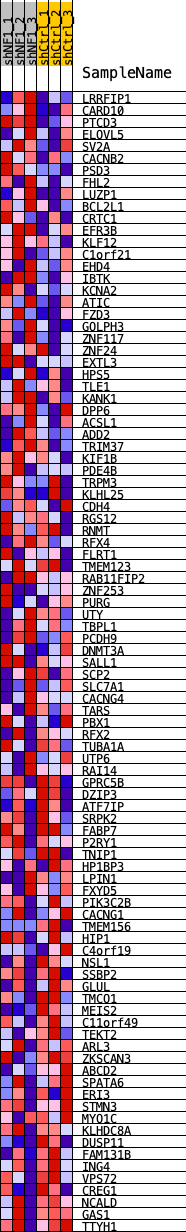
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shNF1 |
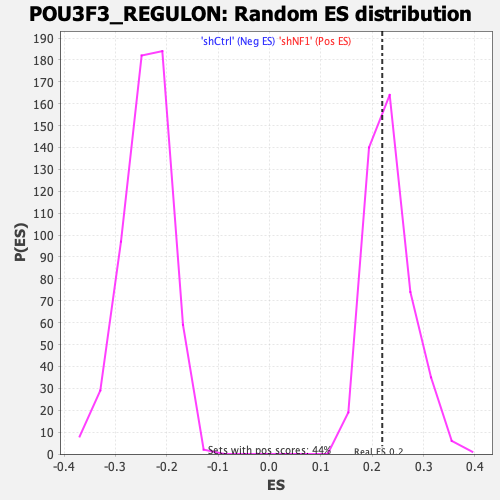
| GeneSet | POU3F3_REGULON |
| Enrichment Score (ES) | 0.21948913 |
| Normalized Enrichment Score (NES) | 0.94285506 |
| Nominal p-value | 0.5808656 |
| FDR q-value | 0.59351146 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LRRFIP1 | LRRFIP1 | 5 | 0.137 | 0.0518 | Yes |
| 2 | CARD10 | CARD10 | 51 | 0.100 | 0.0875 | Yes |
| 3 | PTCD3 | PTCD3 | 150 | 0.083 | 0.1144 | Yes |
| 4 | ELOVL5 | ELOVL5 | 1250 | 0.047 | 0.0810 | Yes |
| 5 | SV2A | SV2A | 1300 | 0.046 | 0.0962 | Yes |
| 6 | CACNB2 | CACNB2 | 1353 | 0.045 | 0.1109 | Yes |
| 7 | PSD3 | PSD3 | 1363 | 0.045 | 0.1276 | Yes |
| 8 | FHL2 | FHL2 | 1493 | 0.043 | 0.1381 | Yes |
| 9 | LUZP1 | LUZP1 | 1593 | 0.043 | 0.1497 | Yes |
| 10 | BCL2L1 | BCL2L1 | 1730 | 0.041 | 0.1590 | Yes |
| 11 | CRTC1 | CRTC1 | 2005 | 0.039 | 0.1609 | Yes |
| 12 | EFR3B | EFR3B | 2305 | 0.036 | 0.1606 | Yes |
| 13 | KLF12 | KLF12 | 2473 | 0.035 | 0.1660 | Yes |
| 14 | C1orf21 | C1orf21 | 2595 | 0.034 | 0.1732 | Yes |
| 15 | EHD4 | EHD4 | 2689 | 0.033 | 0.1815 | Yes |
| 16 | IBTK | IBTK | 3050 | 0.031 | 0.1764 | Yes |
| 17 | KCNA2 | KCNA2 | 3071 | 0.031 | 0.1871 | Yes |
| 18 | ATIC | ATIC | 3081 | 0.030 | 0.1982 | Yes |
| 19 | FZD3 | FZD3 | 3087 | 0.030 | 0.2095 | Yes |
| 20 | GOLPH3 | GOLPH3 | 3145 | 0.030 | 0.2183 | Yes |
| 21 | ZNF117 | ZNF117 | 3565 | 0.027 | 0.2092 | Yes |
| 22 | ZNF24 | ZNF24 | 3604 | 0.027 | 0.2178 | Yes |
| 23 | EXTL3 | EXTL3 | 3782 | 0.026 | 0.2195 | Yes |
| 24 | HPS5 | HPS5 | 4444 | 0.023 | 0.1974 | No |
| 25 | TLE1 | TLE1 | 4766 | 0.021 | 0.1906 | No |
| 26 | KANK1 | KANK1 | 4859 | 0.021 | 0.1942 | No |
| 27 | DPP6 | DPP6 | 5104 | 0.020 | 0.1904 | No |
| 28 | ACSL1 | ACSL1 | 5130 | 0.020 | 0.1967 | No |
| 29 | ADD2 | ADD2 | 5163 | 0.020 | 0.2026 | No |
| 30 | TRIM37 | TRIM37 | 5334 | 0.019 | 0.2018 | No |
| 31 | KIF1B | KIF1B | 5455 | 0.018 | 0.2031 | No |
| 32 | PDE4B | PDE4B | 6042 | 0.016 | 0.1818 | No |
| 33 | TRPM3 | TRPM3 | 6059 | 0.016 | 0.1870 | No |
| 34 | KLHL25 | KLHL25 | 6325 | 0.015 | 0.1802 | No |
| 35 | CDH4 | CDH4 | 6379 | 0.014 | 0.1832 | No |
| 36 | RGS12 | RGS12 | 6724 | 0.013 | 0.1721 | No |
| 37 | RNMT | RNMT | 7133 | 0.012 | 0.1575 | No |
| 38 | RFX4 | RFX4 | 7904 | 0.009 | 0.1251 | No |
| 39 | FLRT1 | FLRT1 | 8146 | 0.008 | 0.1169 | No |
| 40 | TMEM123 | TMEM123 | 8774 | 0.006 | 0.0901 | No |
| 41 | RAB11FIP2 | RAB11FIP2 | 8918 | 0.006 | 0.0855 | No |
| 42 | ZNF253 | ZNF253 | 9103 | 0.005 | 0.0788 | No |
| 43 | PURG | PURG | 9522 | 0.004 | 0.0608 | No |
| 44 | UTY | UTY | 9620 | 0.003 | 0.0575 | No |
| 45 | TBPL1 | TBPL1 | 9781 | 0.003 | 0.0511 | No |
| 46 | PCDH9 | PCDH9 | 9850 | 0.002 | 0.0488 | No |
| 47 | DNMT3A | DNMT3A | 10310 | 0.001 | 0.0279 | No |
| 48 | SALL1 | SALL1 | 10618 | 0.000 | 0.0137 | No |
| 49 | SCP2 | SCP2 | 10700 | -0.000 | 0.0099 | No |
| 50 | SLC7A1 | SLC7A1 | 10948 | -0.001 | -0.0012 | No |
| 51 | CACNG4 | CACNG4 | 11146 | -0.002 | -0.0098 | No |
| 52 | TARS | TARS | 11342 | -0.002 | -0.0180 | No |
| 53 | PBX1 | PBX1 | 11395 | -0.002 | -0.0195 | No |
| 54 | RFX2 | RFX2 | 12049 | -0.005 | -0.0482 | No |
| 55 | TUBA1A | TUBA1A | 12515 | -0.006 | -0.0675 | No |
| 56 | UTP6 | UTP6 | 13423 | -0.009 | -0.1062 | No |
| 57 | RAI14 | RAI14 | 13643 | -0.010 | -0.1127 | No |
| 58 | GPRC5B | GPRC5B | 13715 | -0.010 | -0.1122 | No |
| 59 | DZIP3 | DZIP3 | 13776 | -0.010 | -0.1111 | No |
| 60 | ATF7IP | ATF7IP | 13904 | -0.011 | -0.1129 | No |
| 61 | SRPK2 | SRPK2 | 13946 | -0.011 | -0.1107 | No |
| 62 | FABP7 | FABP7 | 14093 | -0.011 | -0.1132 | No |
| 63 | P2RY1 | P2RY1 | 14374 | -0.012 | -0.1215 | No |
| 64 | TNIP1 | TNIP1 | 14388 | -0.012 | -0.1174 | No |
| 65 | HP1BP3 | HP1BP3 | 14549 | -0.013 | -0.1199 | No |
| 66 | LPIN1 | LPIN1 | 14700 | -0.014 | -0.1217 | No |
| 67 | FXYD5 | FXYD5 | 14924 | -0.014 | -0.1265 | No |
| 68 | PIK3C2B | PIK3C2B | 14984 | -0.015 | -0.1237 | No |
| 69 | CACNG1 | CACNG1 | 15275 | -0.016 | -0.1311 | No |
| 70 | TMEM156 | TMEM156 | 15326 | -0.016 | -0.1273 | No |
| 71 | HIP1 | HIP1 | 15693 | -0.018 | -0.1377 | No |
| 72 | C4orf19 | C4orf19 | 15905 | -0.019 | -0.1404 | No |
| 73 | NSL1 | NSL1 | 16698 | -0.022 | -0.1688 | No |
| 74 | SSBP2 | SSBP2 | 16700 | -0.022 | -0.1604 | No |
| 75 | GLUL | GLUL | 16877 | -0.023 | -0.1598 | No |
| 76 | TMCO1 | TMCO1 | 16914 | -0.023 | -0.1526 | No |
| 77 | MEIS2 | MEIS2 | 17076 | -0.024 | -0.1510 | No |
| 78 | C11orf49 | C11orf49 | 18227 | -0.031 | -0.1929 | No |
| 79 | TEKT2 | TEKT2 | 18606 | -0.033 | -0.1981 | No |
| 80 | ARL3 | ARL3 | 18627 | -0.033 | -0.1865 | No |
| 81 | ZKSCAN3 | ZKSCAN3 | 18909 | -0.035 | -0.1863 | No |
| 82 | ABCD2 | ABCD2 | 18967 | -0.035 | -0.1756 | No |
| 83 | SPATA6 | SPATA6 | 19249 | -0.037 | -0.1744 | No |
| 84 | ERI3 | ERI3 | 19381 | -0.039 | -0.1659 | No |
| 85 | STMN3 | STMN3 | 19531 | -0.040 | -0.1578 | No |
| 86 | MYO1C | MYO1C | 19667 | -0.041 | -0.1485 | No |
| 87 | KLHDC8A | KLHDC8A | 19821 | -0.042 | -0.1395 | No |
| 88 | DUSP11 | DUSP11 | 19870 | -0.043 | -0.1254 | No |
| 89 | FAM131B | FAM131B | 19958 | -0.044 | -0.1127 | No |
| 90 | ING4 | ING4 | 20331 | -0.049 | -0.1115 | No |
| 91 | VPS72 | VPS72 | 20799 | -0.057 | -0.1115 | No |
| 92 | CREG1 | CREG1 | 20852 | -0.058 | -0.0917 | No |
| 93 | NCALD | NCALD | 21072 | -0.065 | -0.0772 | No |
| 94 | GAS1 | GAS1 | 21567 | -0.130 | -0.0507 | No |
| 95 | TTYH1 | TTYH1 | 21573 | -0.137 | 0.0011 | No |