Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
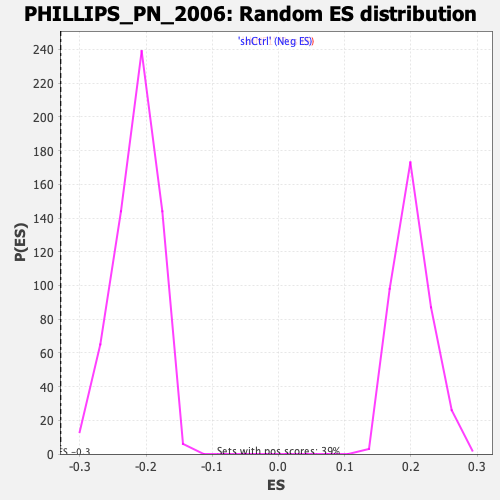
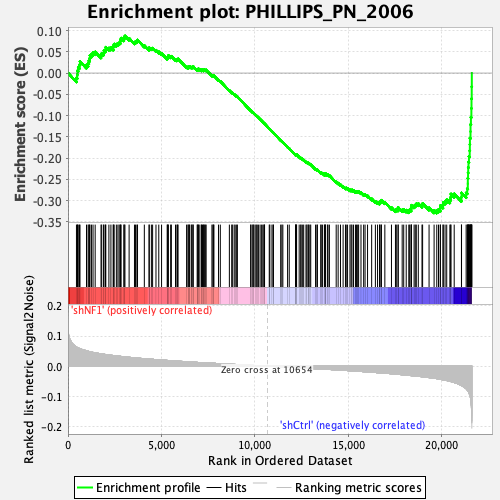
| GeneSet | PHILLIPS_PN_2006 |
| Enrichment Score (ES) | -0.32946315 |
| Normalized Enrichment Score (NES) | -1.5353752 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016571008 |
| FWER p-Value | 0.136 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CNTN1 | CNTN1 | 452 | 0.063 | -0.0112 | No |
| 2 | OPCML | OPCML | 497 | 0.062 | -0.0035 | No |
| 3 | KSR2 | KSR2 | 508 | 0.061 | 0.0057 | No |
| 4 | DNM3 | DNM3 | 548 | 0.060 | 0.0134 | No |
| 5 | TPM1 | TPM1 | 614 | 0.058 | 0.0195 | No |
| 6 | F2 | F2 | 641 | 0.058 | 0.0274 | No |
| 7 | GABRA3 | GABRA3 | 989 | 0.051 | 0.0192 | No |
| 8 | SNAP91 | SNAP91 | 1075 | 0.049 | 0.0230 | No |
| 9 | NUMA1 | NUMA1 | 1116 | 0.049 | 0.0288 | No |
| 10 | DSCAML1 | DSCAML1 | 1156 | 0.048 | 0.0345 | No |
| 11 | PSD | PSD | 1172 | 0.048 | 0.0414 | No |
| 12 | NOG | NOG | 1260 | 0.047 | 0.0446 | No |
| 13 | NKAIN4 | NKAIN4 | 1339 | 0.045 | 0.0482 | No |
| 14 | C12orf53 | C12orf53 | 1453 | 0.044 | 0.0498 | No |
| 15 | SSTR2 | SSTR2 | 1766 | 0.041 | 0.0416 | No |
| 16 | FAM19A5 | FAM19A5 | 1803 | 0.040 | 0.0463 | No |
| 17 | SHD | SHD | 1903 | 0.039 | 0.0479 | No |
| 18 | ARHGAP22 | ARHGAP22 | 1924 | 0.039 | 0.0532 | No |
| 19 | CRTC1 | CRTC1 | 2005 | 0.039 | 0.0555 | No |
| 20 | SEZ6L | SEZ6L | 2022 | 0.038 | 0.0608 | No |
| 21 | FGF14 | FGF14 | 2187 | 0.037 | 0.0590 | No |
| 22 | RAB11FIP4 | RAB11FIP4 | 2295 | 0.036 | 0.0597 | No |
| 23 | NTRK2 | NTRK2 | 2422 | 0.035 | 0.0593 | No |
| 24 | WDR86 | WDR86 | 2423 | 0.035 | 0.0649 | No |
| 25 | RPRM | RPRM | 2469 | 0.035 | 0.0683 | No |
| 26 | SGCG | SGCG | 2586 | 0.034 | 0.0682 | No |
| 27 | SLC25A21 | SLC25A21 | 2669 | 0.033 | 0.0696 | No |
| 28 | RTN1 | RTN1 | 2737 | 0.033 | 0.0716 | No |
| 29 | SLITRK5 | SLITRK5 | 2798 | 0.032 | 0.0739 | No |
| 30 | KCNB1 | KCNB1 | 2803 | 0.032 | 0.0788 | No |
| 31 | OVOL1 | OVOL1 | 2840 | 0.032 | 0.0822 | No |
| 32 | EPB41L2 | EPB41L2 | 2988 | 0.031 | 0.0802 | No |
| 33 | OMG | OMG | 3005 | 0.031 | 0.0844 | No |
| 34 | GLUD2 | GLUD2 | 3045 | 0.031 | 0.0874 | No |
| 35 | ZNF488 | ZNF488 | 3268 | 0.029 | 0.0816 | No |
| 36 | AP2B1 | AP2B1 | 3555 | 0.028 | 0.0726 | No |
| 37 | SLC1A1 | SLC1A1 | 3612 | 0.027 | 0.0743 | No |
| 38 | WNT7B | WNT7B | 3684 | 0.027 | 0.0752 | No |
| 39 | SLC1A4 | SLC1A4 | 3718 | 0.027 | 0.0778 | No |
| 40 | FLJ16779 | FLJ16779 | 4082 | 0.025 | 0.0647 | No |
| 41 | PLCB1 | PLCB1 | 4337 | 0.023 | 0.0565 | No |
| 42 | GRIK4 | GRIK4 | 4343 | 0.023 | 0.0599 | No |
| 43 | psiTPTE22 | psiTPTE22 | 4459 | 0.023 | 0.0581 | No |
| 44 | GABBR2 | GABBR2 | 4525 | 0.022 | 0.0586 | No |
| 45 | CYFIP2 | CYFIP2 | 4705 | 0.022 | 0.0537 | No |
| 46 | HS3ST4 | HS3ST4 | 4855 | 0.021 | 0.0500 | No |
| 47 | FGF12 | FGF12 | 5004 | 0.020 | 0.0463 | No |
| 48 | P2RY13 | P2RY13 | 5304 | 0.019 | 0.0353 | No |
| 49 | AJAP1 | AJAP1 | 5329 | 0.019 | 0.0371 | No |
| 50 | MMP16 | MMP16 | 5348 | 0.019 | 0.0392 | No |
| 51 | FSD1 | FSD1 | 5366 | 0.019 | 0.0413 | No |
| 52 | PHACTR3 | PHACTR3 | 5491 | 0.018 | 0.0384 | No |
| 53 | DSCAM | DSCAM | 5527 | 0.018 | 0.0396 | No |
| 54 | SMOC1 | SMOC1 | 5750 | 0.017 | 0.0318 | No |
| 55 | TMLHE | TMLHE | 5825 | 0.017 | 0.0310 | No |
| 56 | PARD3 | PARD3 | 5860 | 0.016 | 0.0320 | No |
| 57 | ASB13 | ASB13 | 5884 | 0.016 | 0.0335 | No |
| 58 | PRKCZ | PRKCZ | 6342 | 0.014 | 0.0144 | No |
| 59 | ABLIM3 | ABLIM3 | 6425 | 0.014 | 0.0128 | No |
| 60 | NCAM1 | NCAM1 | 6426 | 0.014 | 0.0150 | No |
| 61 | CALCRL | CALCRL | 6485 | 0.014 | 0.0145 | No |
| 62 | HIP1R | HIP1R | 6508 | 0.014 | 0.0156 | No |
| 63 | PAR5 | PAR5 | 6596 | 0.014 | 0.0137 | No |
| 64 | PCSK6 | PCSK6 | 6631 | 0.013 | 0.0142 | No |
| 65 | KCTD4 | KCTD4 | 6687 | 0.013 | 0.0137 | No |
| 66 | FSTL5 | FSTL5 | 6693 | 0.013 | 0.0156 | No |
| 67 | FERMT1 | FERMT1 | 6908 | 0.012 | 0.0075 | No |
| 68 | FAM133A | FAM133A | 6945 | 0.012 | 0.0077 | No |
| 69 | DPP10 | DPP10 | 6951 | 0.012 | 0.0094 | No |
| 70 | GGTA1 | GGTA1 | 6975 | 0.012 | 0.0102 | No |
| 71 | MAPK8IP2 | MAPK8IP2 | 7050 | 0.012 | 0.0086 | No |
| 72 | GNAO1 | GNAO1 | 7134 | 0.012 | 0.0066 | No |
| 73 | MARCH8 | MARCH8 | 7150 | 0.011 | 0.0077 | No |
| 74 | ZC3H12B | ZC3H12B | 7194 | 0.011 | 0.0074 | No |
| 75 | FRY | FRY | 7253 | 0.011 | 0.0065 | No |
| 76 | GFRA1 | GFRA1 | 7266 | 0.011 | 0.0077 | No |
| 77 | C11orf2 | C11orf2 | 7293 | 0.011 | 0.0082 | No |
| 78 | AKR1C1 | AKR1C1 | 7362 | 0.011 | 0.0067 | No |
| 79 | B3GAT1 | B3GAT1 | 7384 | 0.011 | 0.0074 | No |
| 80 | ACSM5 | ACSM5 | 7698 | 0.010 | -0.0058 | No |
| 81 | EFHA2 | EFHA2 | 7752 | 0.009 | -0.0068 | No |
| 82 | SCD | SCD | 7776 | 0.009 | -0.0064 | No |
| 83 | FBXO2 | FBXO2 | 7804 | 0.009 | -0.0062 | No |
| 84 | NTN4 | NTN4 | 8051 | 0.008 | -0.0164 | No |
| 85 | FLRT1 | FLRT1 | 8146 | 0.008 | -0.0195 | No |
| 86 | ZCCHC24 | ZCCHC24 | 8630 | 0.006 | -0.0411 | No |
| 87 | PTGDS | PTGDS | 8758 | 0.006 | -0.0460 | No |
| 88 | KIF21B | KIF21B | 8787 | 0.006 | -0.0464 | No |
| 89 | BMP2 | BMP2 | 8885 | 0.006 | -0.0501 | No |
| 90 | RASL10A | RASL10A | 8941 | 0.006 | -0.0518 | No |
| 91 | RIMS2 | RIMS2 | 9020 | 0.005 | -0.0546 | No |
| 92 | IKZF5 | IKZF5 | 9058 | 0.005 | -0.0555 | No |
| 93 | RIPPLY2 | RIPPLY2 | 9767 | 0.003 | -0.0882 | No |
| 94 | RGS9 | RGS9 | 9789 | 0.003 | -0.0888 | No |
| 95 | GALNT13 | GALNT13 | 9886 | 0.002 | -0.0929 | No |
| 96 | NET1 | NET1 | 9895 | 0.002 | -0.0929 | No |
| 97 | REPS2 | REPS2 | 9937 | 0.002 | -0.0945 | No |
| 98 | KLRC3 | KLRC3 | 10008 | 0.002 | -0.0974 | No |
| 99 | HDAC5 | HDAC5 | 10070 | 0.002 | -0.1000 | No |
| 100 | LRRC4 | LRRC4 | 10121 | 0.002 | -0.1021 | No |
| 101 | PKP4 | PKP4 | 10183 | 0.001 | -0.1047 | No |
| 102 | GPR158 | GPR158 | 10271 | 0.001 | -0.1086 | No |
| 103 | CALN1 | CALN1 | 10342 | 0.001 | -0.1117 | No |
| 104 | CSMD3 | CSMD3 | 10417 | 0.001 | -0.1151 | No |
| 105 | FAM155A | FAM155A | 10474 | 0.001 | -0.1176 | No |
| 106 | ELMO1 | ELMO1 | 10501 | 0.000 | -0.1187 | No |
| 107 | SPHKAP | SPHKAP | 10512 | 0.000 | -0.1191 | No |
| 108 | NAP1L3 | NAP1L3 | 10763 | -0.000 | -0.1308 | No |
| 109 | STOX1 | STOX1 | 10795 | -0.000 | -0.1321 | No |
| 110 | SLIT1 | SLIT1 | 10908 | -0.001 | -0.1372 | No |
| 111 | CBX7 | CBX7 | 10970 | -0.001 | -0.1399 | No |
| 112 | EHD3 | EHD3 | 10988 | -0.001 | -0.1406 | No |
| 113 | TRIM31 | TRIM31 | 11374 | -0.002 | -0.1582 | No |
| 114 | FUT9 | FUT9 | 11437 | -0.003 | -0.1607 | No |
| 115 | FAM110B | FAM110B | 11490 | -0.003 | -0.1627 | No |
| 116 | PKNOX2 | PKNOX2 | 11744 | -0.004 | -0.1740 | No |
| 117 | RPL37 | RPL37 | 11832 | -0.004 | -0.1775 | No |
| 118 | GABBR1 | GABBR1 | 12173 | -0.005 | -0.1926 | No |
| 119 | RASGEF1C | RASGEF1C | 12180 | -0.005 | -0.1921 | No |
| 120 | MAPT | MAPT | 12192 | -0.005 | -0.1918 | No |
| 121 | SORBS1 | SORBS1 | 12199 | -0.005 | -0.1913 | No |
| 122 | DOK6 | DOK6 | 12225 | -0.005 | -0.1917 | No |
| 123 | SORCS3 | SORCS3 | 12363 | -0.006 | -0.1972 | No |
| 124 | ALDH5A1 | ALDH5A1 | 12447 | -0.006 | -0.2001 | No |
| 125 | KIF5A | KIF5A | 12467 | -0.006 | -0.2001 | No |
| 126 | C13orf15 | C13orf15 | 12550 | -0.006 | -0.2029 | No |
| 127 | KCNN3 | KCNN3 | 12605 | -0.006 | -0.2044 | No |
| 128 | TPCN2 | TPCN2 | 12728 | -0.007 | -0.2091 | No |
| 129 | P2RX7 | P2RX7 | 12774 | -0.007 | -0.2101 | No |
| 130 | ANKS1B | ANKS1B | 12848 | -0.007 | -0.2124 | No |
| 131 | THRA | THRA | 12849 | -0.007 | -0.2112 | No |
| 132 | LUZP2 | LUZP2 | 12933 | -0.007 | -0.2140 | No |
| 133 | TMOD2 | TMOD2 | 12981 | -0.008 | -0.2149 | No |
| 134 | SMAD9 | SMAD9 | 13239 | -0.008 | -0.2256 | No |
| 135 | NRG3 | NRG3 | 13312 | -0.009 | -0.2276 | No |
| 136 | ATRNL1 | ATRNL1 | 13338 | -0.009 | -0.2274 | No |
| 137 | HSPA12A | HSPA12A | 13516 | -0.009 | -0.2342 | No |
| 138 | NRSN1 | NRSN1 | 13542 | -0.009 | -0.2339 | No |
| 139 | SEC31B | SEC31B | 13615 | -0.010 | -0.2357 | No |
| 140 | GPRC5B | GPRC5B | 13715 | -0.010 | -0.2388 | No |
| 141 | SUSD5 | SUSD5 | 13751 | -0.010 | -0.2388 | No |
| 142 | CSDC2 | CSDC2 | 13756 | -0.010 | -0.2374 | No |
| 143 | GRIA1 | GRIA1 | 13757 | -0.010 | -0.2358 | No |
| 144 | NDRG2 | NDRG2 | 13872 | -0.011 | -0.2395 | No |
| 145 | GNAL | GNAL | 13906 | -0.011 | -0.2393 | No |
| 146 | SCN3A | SCN3A | 13980 | -0.011 | -0.2410 | No |
| 147 | TNKS2 | TNKS2 | 14343 | -0.012 | -0.2560 | No |
| 148 | MN1 | MN1 | 14447 | -0.013 | -0.2588 | No |
| 149 | SSTR1 | SSTR1 | 14573 | -0.013 | -0.2626 | No |
| 150 | RAC3 | RAC3 | 14713 | -0.014 | -0.2669 | No |
| 151 | LGR5 | LGR5 | 14839 | -0.014 | -0.2705 | No |
| 152 | GRIA4 | GRIA4 | 14879 | -0.014 | -0.2701 | No |
| 153 | ABHD6 | ABHD6 | 14951 | -0.015 | -0.2711 | No |
| 154 | SOX6 | SOX6 | 15079 | -0.015 | -0.2747 | No |
| 155 | JPH4 | JPH4 | 15142 | -0.015 | -0.2751 | No |
| 156 | APOE | APOE | 15188 | -0.016 | -0.2748 | No |
| 157 | USH1C | USH1C | 15270 | -0.016 | -0.2761 | No |
| 158 | GABRB3 | GABRB3 | 15373 | -0.016 | -0.2783 | No |
| 159 | ZNF804A | ZNF804A | 15432 | -0.017 | -0.2784 | No |
| 160 | SGSM1 | SGSM1 | 15492 | -0.017 | -0.2785 | No |
| 161 | CNTN3 | CNTN3 | 15542 | -0.017 | -0.2781 | No |
| 162 | BEND7 | BEND7 | 15669 | -0.018 | -0.2812 | No |
| 163 | EPHB1 | EPHB1 | 15812 | -0.018 | -0.2850 | No |
| 164 | FGF13 | FGF13 | 15887 | -0.019 | -0.2855 | No |
| 165 | GRID1 | GRID1 | 16020 | -0.019 | -0.2887 | No |
| 166 | CDR1 | CDR1 | 16240 | -0.020 | -0.2957 | No |
| 167 | ABLIM1 | ABLIM1 | 16439 | -0.021 | -0.3017 | No |
| 168 | PDK4 | PDK4 | 16555 | -0.022 | -0.3037 | No |
| 169 | CCNK | CCNK | 16660 | -0.022 | -0.3050 | No |
| 170 | GLUD1 | GLUD1 | 16679 | -0.022 | -0.3024 | No |
| 171 | DNAJC12 | DNAJC12 | 16742 | -0.022 | -0.3017 | No |
| 172 | ID4 | ID4 | 16762 | -0.023 | -0.2990 | No |
| 173 | MPPE1 | MPPE1 | 16945 | -0.023 | -0.3039 | No |
| 174 | JPH3 | JPH3 | 17299 | -0.025 | -0.3164 | No |
| 175 | SLITRK2 | SLITRK2 | 17512 | -0.026 | -0.3222 | No |
| 176 | TMEM100 | TMEM100 | 17557 | -0.027 | -0.3200 | No |
| 177 | NALCN | NALCN | 17655 | -0.027 | -0.3203 | No |
| 178 | MCF2 | MCF2 | 17656 | -0.027 | -0.3160 | No |
| 179 | RASSF4 | RASSF4 | 17872 | -0.028 | -0.3216 | No |
| 180 | PLEKHB1 | PLEKHB1 | 17954 | -0.029 | -0.3208 | No |
| 181 | ADCY2 | ADCY2 | 18088 | -0.030 | -0.3223 | No |
| 182 | CECR6 | CECR6 | 18226 | -0.031 | -0.3239 | Yes |
| 183 | TAL1 | TAL1 | 18273 | -0.031 | -0.3212 | Yes |
| 184 | DUSP26 | DUSP26 | 18351 | -0.031 | -0.3199 | Yes |
| 185 | KCNQ5 | KCNQ5 | 18355 | -0.031 | -0.3151 | Yes |
| 186 | PDE2A | PDE2A | 18370 | -0.031 | -0.3108 | Yes |
| 187 | PDK2 | PDK2 | 18509 | -0.032 | -0.3121 | Yes |
| 188 | HLF | HLF | 18571 | -0.033 | -0.3099 | Yes |
| 189 | ARL3 | ARL3 | 18627 | -0.033 | -0.3072 | Yes |
| 190 | ZDHHC22 | ZDHHC22 | 18734 | -0.034 | -0.3069 | Yes |
| 191 | FAIM2 | FAIM2 | 18932 | -0.035 | -0.3106 | Yes |
| 192 | GAB2 | GAB2 | 18965 | -0.035 | -0.3065 | Yes |
| 193 | RPL13 | RPL13 | 19310 | -0.038 | -0.3166 | Yes |
| 194 | DTX4 | DTX4 | 19579 | -0.040 | -0.3228 | Yes |
| 195 | GAD1 | GAD1 | 19722 | -0.041 | -0.3229 | Yes |
| 196 | IL17D | IL17D | 19815 | -0.042 | -0.3206 | Yes |
| 197 | CMTM5 | CMTM5 | 19908 | -0.044 | -0.3180 | Yes |
| 198 | C1QL1 | C1QL1 | 19916 | -0.044 | -0.3114 | Yes |
| 199 | TMEM59L | TMEM59L | 20054 | -0.045 | -0.3107 | Yes |
| 200 | RUNDC3A | RUNDC3A | 20069 | -0.045 | -0.3042 | Yes |
| 201 | ATP6V1G2 | ATP6V1G2 | 20165 | -0.046 | -0.3013 | Yes |
| 202 | BCAN | BCAN | 20254 | -0.048 | -0.2979 | Yes |
| 203 | SNRPN | SNRPN | 20423 | -0.050 | -0.2979 | Yes |
| 204 | MAF | MAF | 20468 | -0.051 | -0.2919 | Yes |
| 205 | GRIA2 | GRIA2 | 20478 | -0.051 | -0.2843 | Yes |
| 206 | PID1 | PID1 | 20654 | -0.054 | -0.2839 | Yes |
| 207 | FBXL15 | FBXL15 | 21042 | -0.064 | -0.2919 | Yes |
| 208 | ALDOC | ALDOC | 21046 | -0.064 | -0.2819 | Yes |
| 209 | OLIG2 | OLIG2 | 21293 | -0.075 | -0.2816 | Yes |
| 210 | DLL1 | DLL1 | 21352 | -0.079 | -0.2718 | Yes |
| 211 | ENHO | ENHO | 21382 | -0.082 | -0.2602 | Yes |
| 212 | PHYHIPL | PHYHIPL | 21383 | -0.082 | -0.2473 | Yes |
| 213 | PCSK1N | PCSK1N | 21396 | -0.083 | -0.2348 | Yes |
| 214 | FXYD6 | FXYD6 | 21404 | -0.084 | -0.2219 | Yes |
| 215 | SH3GL2 | SH3GL2 | 21419 | -0.085 | -0.2091 | Yes |
| 216 | CRYAB | CRYAB | 21443 | -0.089 | -0.1961 | Yes |
| 217 | SCG3 | SCG3 | 21484 | -0.096 | -0.1829 | Yes |
| 218 | HEY2 | HEY2 | 21491 | -0.096 | -0.1680 | Yes |
| 219 | KIF1A | KIF1A | 21496 | -0.097 | -0.1528 | Yes |
| 220 | DLL3 | DLL3 | 21526 | -0.105 | -0.1376 | Yes |
| 221 | OLIG1 | OLIG1 | 21528 | -0.106 | -0.1209 | Yes |
| 222 | NEU4 | NEU4 | 21549 | -0.115 | -0.1037 | Yes |
| 223 | TTYH1 | TTYH1 | 21573 | -0.137 | -0.0831 | Yes |
| 224 | TIMP4 | TIMP4 | 21580 | -0.147 | -0.0603 | Yes |
| 225 | SOX8 | SOX8 | 21591 | -0.180 | -0.0323 | Yes |
| 226 | ASCL1 | ASCL1 | 21596 | -0.206 | -0.0000 | Yes |