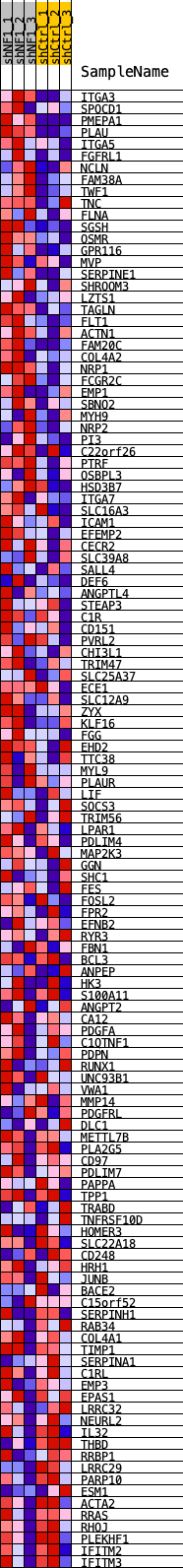
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shNF1 |
| GeneSet | PHILLIPS_MES_2006 |
| Enrichment Score (ES) | 0.41752848 |
| Normalized Enrichment Score (NES) | 1.8970993 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.0769233E-4 |
| FWER p-Value | 0.001 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ITGA3 | ITGA3 | 22 | 0.116 | 0.0299 | Yes |
| 2 | SPOCD1 | SPOCD1 | 79 | 0.093 | 0.0520 | Yes |
| 3 | PMEPA1 | PMEPA1 | 89 | 0.091 | 0.0760 | Yes |
| 4 | PLAU | PLAU | 94 | 0.090 | 0.0999 | Yes |
| 5 | ITGA5 | ITGA5 | 208 | 0.077 | 0.1151 | Yes |
| 6 | FGFRL1 | FGFRL1 | 273 | 0.072 | 0.1313 | Yes |
| 7 | NCLN | NCLN | 282 | 0.072 | 0.1501 | Yes |
| 8 | FAM38A | FAM38A | 339 | 0.068 | 0.1657 | Yes |
| 9 | TWF1 | TWF1 | 483 | 0.062 | 0.1756 | Yes |
| 10 | TNC | TNC | 488 | 0.062 | 0.1920 | Yes |
| 11 | FLNA | FLNA | 493 | 0.062 | 0.2083 | Yes |
| 12 | SGSH | SGSH | 554 | 0.060 | 0.2215 | Yes |
| 13 | OSMR | OSMR | 580 | 0.059 | 0.2361 | Yes |
| 14 | GPR116 | GPR116 | 667 | 0.057 | 0.2473 | Yes |
| 15 | MVP | MVP | 711 | 0.056 | 0.2603 | Yes |
| 16 | SERPINE1 | SERPINE1 | 725 | 0.056 | 0.2746 | Yes |
| 17 | SHROOM3 | SHROOM3 | 797 | 0.055 | 0.2859 | Yes |
| 18 | LZTS1 | LZTS1 | 954 | 0.051 | 0.2923 | Yes |
| 19 | TAGLN | TAGLN | 956 | 0.051 | 0.3060 | Yes |
| 20 | FLT1 | FLT1 | 1023 | 0.050 | 0.3163 | Yes |
| 21 | ACTN1 | ACTN1 | 1227 | 0.047 | 0.3193 | Yes |
| 22 | FAM20C | FAM20C | 1251 | 0.047 | 0.3307 | Yes |
| 23 | COL4A2 | COL4A2 | 1397 | 0.045 | 0.3359 | Yes |
| 24 | NRP1 | NRP1 | 1414 | 0.044 | 0.3470 | Yes |
| 25 | FCGR2C | FCGR2C | 1445 | 0.044 | 0.3573 | Yes |
| 26 | EMP1 | EMP1 | 1560 | 0.043 | 0.3635 | Yes |
| 27 | SBNO2 | SBNO2 | 1719 | 0.041 | 0.3671 | Yes |
| 28 | MYH9 | MYH9 | 1778 | 0.041 | 0.3753 | Yes |
| 29 | NRP2 | NRP2 | 1810 | 0.040 | 0.3846 | Yes |
| 30 | PI3 | PI3 | 1922 | 0.039 | 0.3899 | Yes |
| 31 | C22orf26 | C22orf26 | 2035 | 0.038 | 0.3949 | Yes |
| 32 | PTRF | PTRF | 2260 | 0.036 | 0.3942 | Yes |
| 33 | OSBPL3 | OSBPL3 | 2755 | 0.033 | 0.3799 | Yes |
| 34 | HSD3B7 | HSD3B7 | 2762 | 0.033 | 0.3883 | Yes |
| 35 | ITGA7 | ITGA7 | 2900 | 0.032 | 0.3904 | Yes |
| 36 | SLC16A3 | SLC16A3 | 2914 | 0.032 | 0.3982 | Yes |
| 37 | ICAM1 | ICAM1 | 2950 | 0.031 | 0.4049 | Yes |
| 38 | EFEMP2 | EFEMP2 | 3022 | 0.031 | 0.4098 | Yes |
| 39 | CECR2 | CECR2 | 3122 | 0.030 | 0.4133 | Yes |
| 40 | SLC39A8 | SLC39A8 | 3202 | 0.030 | 0.4175 | Yes |
| 41 | SALL4 | SALL4 | 3654 | 0.027 | 0.4037 | No |
| 42 | DEF6 | DEF6 | 3658 | 0.027 | 0.4107 | No |
| 43 | ANGPTL4 | ANGPTL4 | 3926 | 0.025 | 0.4051 | No |
| 44 | STEAP3 | STEAP3 | 3930 | 0.025 | 0.4117 | No |
| 45 | C1R | C1R | 4214 | 0.024 | 0.4049 | No |
| 46 | CD151 | CD151 | 4233 | 0.024 | 0.4104 | No |
| 47 | PVRL2 | PVRL2 | 4592 | 0.022 | 0.3997 | No |
| 48 | CHI3L1 | CHI3L1 | 4752 | 0.021 | 0.3980 | No |
| 49 | TRIM47 | TRIM47 | 4821 | 0.021 | 0.4004 | No |
| 50 | SLC25A37 | SLC25A37 | 4833 | 0.021 | 0.4055 | No |
| 51 | ECE1 | ECE1 | 5192 | 0.019 | 0.3940 | No |
| 52 | SLC12A9 | SLC12A9 | 5719 | 0.017 | 0.3741 | No |
| 53 | ZYX | ZYX | 5835 | 0.017 | 0.3731 | No |
| 54 | KLF16 | KLF16 | 6291 | 0.015 | 0.3559 | No |
| 55 | FGG | FGG | 6301 | 0.015 | 0.3593 | No |
| 56 | EHD2 | EHD2 | 6388 | 0.014 | 0.3591 | No |
| 57 | TTC38 | TTC38 | 6673 | 0.013 | 0.3494 | No |
| 58 | MYL9 | MYL9 | 6787 | 0.013 | 0.3476 | No |
| 59 | PLAUR | PLAUR | 7477 | 0.010 | 0.3183 | No |
| 60 | LIF | LIF | 7552 | 0.010 | 0.3175 | No |
| 61 | SOCS3 | SOCS3 | 7565 | 0.010 | 0.3196 | No |
| 62 | TRIM56 | TRIM56 | 7615 | 0.010 | 0.3200 | No |
| 63 | LPAR1 | LPAR1 | 7876 | 0.009 | 0.3103 | No |
| 64 | PDLIM4 | PDLIM4 | 7895 | 0.009 | 0.3118 | No |
| 65 | MAP2K3 | MAP2K3 | 8023 | 0.008 | 0.3082 | No |
| 66 | GGN | GGN | 8037 | 0.008 | 0.3098 | No |
| 67 | SHC1 | SHC1 | 8045 | 0.008 | 0.3117 | No |
| 68 | FES | FES | 8143 | 0.008 | 0.3093 | No |
| 69 | FOSL2 | FOSL2 | 8254 | 0.008 | 0.3063 | No |
| 70 | FPR2 | FPR2 | 9452 | 0.004 | 0.2515 | No |
| 71 | EFNB2 | EFNB2 | 9681 | 0.003 | 0.2417 | No |
| 72 | RYR3 | RYR3 | 9730 | 0.003 | 0.2403 | No |
| 73 | FBN1 | FBN1 | 9820 | 0.003 | 0.2368 | No |
| 74 | BCL3 | BCL3 | 9852 | 0.002 | 0.2360 | No |
| 75 | ANPEP | ANPEP | 9908 | 0.002 | 0.2340 | No |
| 76 | HK3 | HK3 | 9971 | 0.002 | 0.2317 | No |
| 77 | S100A11 | S100A11 | 10703 | -0.000 | 0.1977 | No |
| 78 | ANGPT2 | ANGPT2 | 10813 | -0.001 | 0.1928 | No |
| 79 | CA12 | CA12 | 10917 | -0.001 | 0.1882 | No |
| 80 | PDGFA | PDGFA | 11928 | -0.004 | 0.1423 | No |
| 81 | C1QTNF1 | C1QTNF1 | 11992 | -0.004 | 0.1405 | No |
| 82 | PDPN | PDPN | 11998 | -0.004 | 0.1414 | No |
| 83 | RUNX1 | RUNX1 | 12042 | -0.005 | 0.1406 | No |
| 84 | UNC93B1 | UNC93B1 | 12108 | -0.005 | 0.1389 | No |
| 85 | VWA1 | VWA1 | 12201 | -0.005 | 0.1359 | No |
| 86 | MMP14 | MMP14 | 12245 | -0.005 | 0.1353 | No |
| 87 | PDGFRL | PDGFRL | 13079 | -0.008 | 0.0986 | No |
| 88 | DLC1 | DLC1 | 13283 | -0.009 | 0.0915 | No |
| 89 | METTL7B | METTL7B | 14209 | -0.012 | 0.0515 | No |
| 90 | PLA2G5 | PLA2G5 | 14303 | -0.012 | 0.0504 | No |
| 91 | CD97 | CD97 | 14451 | -0.013 | 0.0470 | No |
| 92 | PDLIM7 | PDLIM7 | 14524 | -0.013 | 0.0471 | No |
| 93 | PAPPA | PAPPA | 14536 | -0.013 | 0.0500 | No |
| 94 | TPP1 | TPP1 | 14556 | -0.013 | 0.0526 | No |
| 95 | TRABD | TRABD | 14647 | -0.013 | 0.0520 | No |
| 96 | TNFRSF10D | TNFRSF10D | 14712 | -0.014 | 0.0527 | No |
| 97 | HOMER3 | HOMER3 | 14990 | -0.015 | 0.0438 | No |
| 98 | SLC22A18 | SLC22A18 | 15179 | -0.016 | 0.0392 | No |
| 99 | CD248 | CD248 | 15184 | -0.016 | 0.0431 | No |
| 100 | HRH1 | HRH1 | 15226 | -0.016 | 0.0454 | No |
| 101 | JUNB | JUNB | 15319 | -0.016 | 0.0454 | No |
| 102 | BACE2 | BACE2 | 15352 | -0.016 | 0.0483 | No |
| 103 | C15orf52 | C15orf52 | 15366 | -0.016 | 0.0520 | No |
| 104 | SERPINH1 | SERPINH1 | 15559 | -0.017 | 0.0476 | No |
| 105 | RAB34 | RAB34 | 15944 | -0.019 | 0.0347 | No |
| 106 | COL4A1 | COL4A1 | 16019 | -0.019 | 0.0364 | No |
| 107 | TIMP1 | TIMP1 | 16033 | -0.019 | 0.0409 | No |
| 108 | SERPINA1 | SERPINA1 | 16362 | -0.021 | 0.0312 | No |
| 109 | C1RL | C1RL | 16390 | -0.021 | 0.0355 | No |
| 110 | EMP3 | EMP3 | 16399 | -0.021 | 0.0406 | No |
| 111 | EPAS1 | EPAS1 | 16865 | -0.023 | 0.0251 | No |
| 112 | LRRC32 | LRRC32 | 18378 | -0.031 | -0.0369 | No |
| 113 | NEURL2 | NEURL2 | 19294 | -0.038 | -0.0694 | No |
| 114 | IL32 | IL32 | 19484 | -0.039 | -0.0677 | No |
| 115 | THBD | THBD | 19532 | -0.040 | -0.0593 | No |
| 116 | RRBP1 | RRBP1 | 19589 | -0.040 | -0.0512 | No |
| 117 | LRRC29 | LRRC29 | 20012 | -0.045 | -0.0589 | No |
| 118 | PARP10 | PARP10 | 20078 | -0.045 | -0.0498 | No |
| 119 | ESM1 | ESM1 | 20127 | -0.046 | -0.0398 | No |
| 120 | ACTA2 | ACTA2 | 20255 | -0.048 | -0.0330 | No |
| 121 | RRAS | RRAS | 20323 | -0.049 | -0.0232 | No |
| 122 | RHOJ | RHOJ | 20420 | -0.050 | -0.0143 | No |
| 123 | PLEKHF1 | PLEKHF1 | 21123 | -0.067 | -0.0290 | No |
| 124 | IFITM2 | IFITM2 | 21470 | -0.093 | -0.0203 | No |
| 125 | IFITM3 | IFITM3 | 21498 | -0.098 | 0.0046 | No |