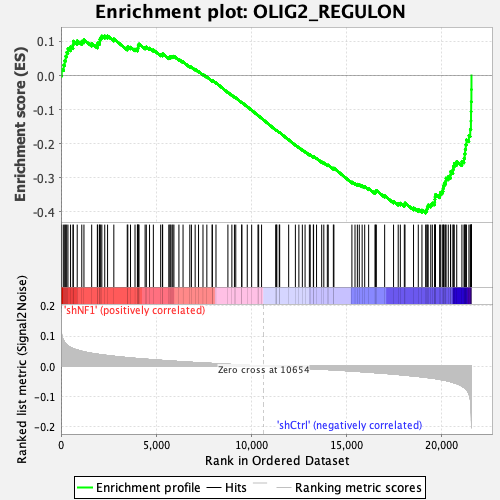
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | OLIG2_REGULON |
| Enrichment Score (ES) | -0.40399104 |
| Normalized Enrichment Score (NES) | -1.7801429 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0018993549 |
| FWER p-Value | 0.011 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | BNC2 | BNC2 | 40 | 0.102 | 0.0186 | No |
| 2 | CD44 | CD44 | 141 | 0.084 | 0.0308 | No |
| 3 | NFAT5 | NFAT5 | 196 | 0.078 | 0.0439 | No |
| 4 | MYOF | MYOF | 245 | 0.074 | 0.0566 | No |
| 5 | AKAP12 | AKAP12 | 299 | 0.070 | 0.0682 | No |
| 6 | ANXA2P1 | ANXA2P1 | 365 | 0.067 | 0.0786 | No |
| 7 | ZNF544 | ZNF544 | 502 | 0.061 | 0.0846 | No |
| 8 | ELL2 | ELL2 | 633 | 0.058 | 0.0901 | No |
| 9 | CGREF1 | CGREF1 | 645 | 0.057 | 0.1011 | No |
| 10 | FSTL1 | FSTL1 | 853 | 0.053 | 0.1022 | No |
| 11 | GCNT1 | GCNT1 | 1093 | 0.049 | 0.1009 | No |
| 12 | PTGER4 | PTGER4 | 1207 | 0.047 | 0.1051 | No |
| 13 | RASSF2 | RASSF2 | 1614 | 0.042 | 0.0946 | No |
| 14 | RASL12 | RASL12 | 1912 | 0.039 | 0.0887 | No |
| 15 | SLIT2 | SLIT2 | 1932 | 0.039 | 0.0957 | No |
| 16 | STARD7 | STARD7 | 2033 | 0.038 | 0.0987 | No |
| 17 | CHST11 | CHST11 | 2038 | 0.038 | 0.1062 | No |
| 18 | FZD7 | FZD7 | 2089 | 0.038 | 0.1114 | No |
| 19 | SLC38A3 | SLC38A3 | 2149 | 0.037 | 0.1161 | No |
| 20 | LGI2 | LGI2 | 2301 | 0.036 | 0.1163 | No |
| 21 | DUSP6 | DUSP6 | 2450 | 0.035 | 0.1164 | No |
| 22 | ATP9B | ATP9B | 2779 | 0.032 | 0.1077 | No |
| 23 | BMP2K | BMP2K | 3485 | 0.028 | 0.0804 | No |
| 24 | SOX11 | SOX11 | 3515 | 0.028 | 0.0846 | No |
| 25 | MAP2K5 | MAP2K5 | 3657 | 0.027 | 0.0834 | No |
| 26 | CAST | CAST | 3894 | 0.026 | 0.0775 | No |
| 27 | IDH1 | IDH1 | 4020 | 0.025 | 0.0767 | No |
| 28 | COL7A1 | COL7A1 | 4041 | 0.025 | 0.0807 | No |
| 29 | LEPROTL1 | LEPROTL1 | 4058 | 0.025 | 0.0850 | No |
| 30 | FSCN1 | FSCN1 | 4059 | 0.025 | 0.0899 | No |
| 31 | APBB2 | APBB2 | 4105 | 0.024 | 0.0927 | No |
| 32 | RAF1 | RAF1 | 4425 | 0.023 | 0.0825 | No |
| 33 | PHACTR2 | PHACTR2 | 4488 | 0.023 | 0.0841 | No |
| 34 | ETV5 | ETV5 | 4658 | 0.022 | 0.0806 | No |
| 35 | KANK1 | KANK1 | 4859 | 0.021 | 0.0755 | No |
| 36 | GNAZ | GNAZ | 5244 | 0.019 | 0.0614 | No |
| 37 | IL1A | IL1A | 5336 | 0.019 | 0.0609 | No |
| 38 | MMP16 | MMP16 | 5348 | 0.019 | 0.0642 | No |
| 39 | RBM38 | RBM38 | 5672 | 0.017 | 0.0525 | No |
| 40 | FRMPD1 | FRMPD1 | 5733 | 0.017 | 0.0532 | No |
| 41 | SNX27 | SNX27 | 5745 | 0.017 | 0.0560 | No |
| 42 | ZYX | ZYX | 5835 | 0.017 | 0.0552 | No |
| 43 | DNAH9 | DNAH9 | 5883 | 0.016 | 0.0563 | No |
| 44 | RRAS2 | RRAS2 | 5941 | 0.016 | 0.0569 | No |
| 45 | RAB27A | RAB27A | 6204 | 0.015 | 0.0477 | No |
| 46 | LMF1 | LMF1 | 6421 | 0.014 | 0.0404 | No |
| 47 | ART3 | ART3 | 6777 | 0.013 | 0.0265 | No |
| 48 | TRIB2 | TRIB2 | 6861 | 0.013 | 0.0251 | No |
| 49 | P4HA2 | P4HA2 | 7058 | 0.012 | 0.0183 | No |
| 50 | GPR68 | GPR68 | 7236 | 0.011 | 0.0123 | No |
| 51 | AGPAT5 | AGPAT5 | 7470 | 0.010 | 0.0035 | No |
| 52 | CYP1B1 | CYP1B1 | 7673 | 0.010 | -0.0039 | No |
| 53 | PIK3R2 | PIK3R2 | 7951 | 0.009 | -0.0151 | No |
| 54 | HOMER1 | HOMER1 | 7963 | 0.009 | -0.0139 | No |
| 55 | PRDM8 | PRDM8 | 8154 | 0.008 | -0.0211 | No |
| 56 | NES | NES | 8781 | 0.006 | -0.0491 | No |
| 57 | SDC2 | SDC2 | 8990 | 0.005 | -0.0577 | No |
| 58 | ABAT | ABAT | 9125 | 0.005 | -0.0630 | No |
| 59 | IRS1 | IRS1 | 9195 | 0.005 | -0.0653 | No |
| 60 | CCND2 | CCND2 | 9507 | 0.004 | -0.0791 | No |
| 61 | BCAR3 | BCAR3 | 9513 | 0.004 | -0.0786 | No |
| 62 | GPD1L | GPD1L | 9799 | 0.003 | -0.0913 | No |
| 63 | GOLM1 | GOLM1 | 10031 | 0.002 | -0.1017 | No |
| 64 | SRGN | SRGN | 10364 | 0.001 | -0.1170 | No |
| 65 | GLDC | GLDC | 10399 | 0.001 | -0.1184 | No |
| 66 | ATXN10 | ATXN10 | 10552 | 0.000 | -0.1255 | No |
| 67 | TRIM24 | TRIM24 | 11296 | -0.002 | -0.1597 | No |
| 68 | TMPRSS5 | TMPRSS5 | 11331 | -0.002 | -0.1608 | No |
| 69 | PTGS2 | PTGS2 | 11356 | -0.002 | -0.1615 | No |
| 70 | VEGFC | VEGFC | 11483 | -0.003 | -0.1668 | No |
| 71 | TBKBP1 | TBKBP1 | 11505 | -0.003 | -0.1673 | No |
| 72 | RGS20 | RGS20 | 11978 | -0.004 | -0.1884 | No |
| 73 | FAM114A1 | FAM114A1 | 12331 | -0.006 | -0.2037 | No |
| 74 | TUBA1A | TUBA1A | 12515 | -0.006 | -0.2110 | No |
| 75 | MFAP2 | MFAP2 | 12698 | -0.007 | -0.2182 | No |
| 76 | NINJ2 | NINJ2 | 12841 | -0.007 | -0.2234 | No |
| 77 | DBI | DBI | 13067 | -0.008 | -0.2323 | No |
| 78 | IFFO1 | IFFO1 | 13112 | -0.008 | -0.2327 | No |
| 79 | TUBB2B | TUBB2B | 13277 | -0.009 | -0.2386 | No |
| 80 | LAPTM4B | LAPTM4B | 13289 | -0.009 | -0.2374 | No |
| 81 | SLC19A3 | SLC19A3 | 13445 | -0.009 | -0.2428 | No |
| 82 | GPRC5B | GPRC5B | 13715 | -0.010 | -0.2533 | No |
| 83 | MAP3K1 | MAP3K1 | 13827 | -0.010 | -0.2564 | No |
| 84 | MAP2 | MAP2 | 14012 | -0.011 | -0.2628 | No |
| 85 | FEZ1 | FEZ1 | 14059 | -0.011 | -0.2627 | No |
| 86 | GAB1 | GAB1 | 14329 | -0.012 | -0.2728 | No |
| 87 | POLR2F | POLR2F | 14358 | -0.012 | -0.2716 | No |
| 88 | VASH1 | VASH1 | 15305 | -0.016 | -0.3125 | No |
| 89 | NME4 | NME4 | 15468 | -0.017 | -0.3167 | No |
| 90 | ATP6V0E2 | ATP6V0E2 | 15592 | -0.017 | -0.3190 | No |
| 91 | HIP1 | HIP1 | 15693 | -0.018 | -0.3201 | No |
| 92 | TRAF4 | TRAF4 | 15844 | -0.018 | -0.3234 | No |
| 93 | RLBP1 | RLBP1 | 15978 | -0.019 | -0.3258 | No |
| 94 | BCHE | BCHE | 16186 | -0.020 | -0.3314 | No |
| 95 | PHLPP1 | PHLPP1 | 16520 | -0.021 | -0.3427 | No |
| 96 | TBC1D22B | TBC1D22B | 16550 | -0.022 | -0.3397 | No |
| 97 | MARCKSL1 | MARCKSL1 | 16590 | -0.022 | -0.3371 | No |
| 98 | ARC | ARC | 17028 | -0.024 | -0.3527 | No |
| 99 | QDPR | QDPR | 17496 | -0.026 | -0.3692 | No |
| 100 | PICK1 | PICK1 | 17741 | -0.028 | -0.3751 | No |
| 101 | ENDOG | ENDOG | 17853 | -0.028 | -0.3745 | No |
| 102 | COL4A6 | COL4A6 | 18063 | -0.030 | -0.3784 | No |
| 103 | SHC3 | SHC3 | 18098 | -0.030 | -0.3740 | No |
| 104 | NDE1 | NDE1 | 18547 | -0.032 | -0.3883 | No |
| 105 | PTPRZ1 | PTPRZ1 | 18797 | -0.034 | -0.3931 | No |
| 106 | PODXL2 | PODXL2 | 18990 | -0.035 | -0.3949 | No |
| 107 | VGLL4 | VGLL4 | 19185 | -0.037 | -0.3966 | Yes |
| 108 | SPATA6 | SPATA6 | 19249 | -0.037 | -0.3920 | Yes |
| 109 | BAALC | BAALC | 19268 | -0.038 | -0.3853 | Yes |
| 110 | TMEM121 | TMEM121 | 19317 | -0.038 | -0.3799 | Yes |
| 111 | CSPG4 | CSPG4 | 19454 | -0.039 | -0.3784 | Yes |
| 112 | ST3GAL6 | ST3GAL6 | 19536 | -0.040 | -0.3742 | Yes |
| 113 | RBBP9 | RBBP9 | 19660 | -0.041 | -0.3718 | Yes |
| 114 | MYO1C | MYO1C | 19667 | -0.041 | -0.3638 | Yes |
| 115 | SLC10A3 | SLC10A3 | 19679 | -0.041 | -0.3561 | Yes |
| 116 | TMEM206 | TMEM206 | 19703 | -0.041 | -0.3489 | Yes |
| 117 | HES1 | HES1 | 19921 | -0.044 | -0.3503 | Yes |
| 118 | FAM131B | FAM131B | 19958 | -0.044 | -0.3431 | Yes |
| 119 | PPFIBP2 | PPFIBP2 | 20055 | -0.045 | -0.3385 | Yes |
| 120 | RAPGEF4 | RAPGEF4 | 20103 | -0.046 | -0.3315 | Yes |
| 121 | CITED1 | CITED1 | 20122 | -0.046 | -0.3232 | Yes |
| 122 | NLGN3 | NLGN3 | 20159 | -0.046 | -0.3155 | Yes |
| 123 | SYT11 | SYT11 | 20241 | -0.047 | -0.3098 | Yes |
| 124 | BCAN | BCAN | 20254 | -0.048 | -0.3008 | Yes |
| 125 | PNMA2 | PNMA2 | 20375 | -0.049 | -0.2965 | Yes |
| 126 | SYT17 | SYT17 | 20494 | -0.051 | -0.2917 | Yes |
| 127 | CREB5 | CREB5 | 20498 | -0.051 | -0.2816 | Yes |
| 128 | NOVA1 | NOVA1 | 20622 | -0.054 | -0.2765 | Yes |
| 129 | HLA-DPA1 | HLA-DPA1 | 20639 | -0.054 | -0.2664 | Yes |
| 130 | IRF1 | IRF1 | 20690 | -0.055 | -0.2577 | Yes |
| 131 | GNG7 | GNG7 | 20820 | -0.058 | -0.2521 | Yes |
| 132 | S100B | S100B | 21093 | -0.065 | -0.2516 | Yes |
| 133 | CCDC28B | CCDC28B | 21203 | -0.070 | -0.2426 | Yes |
| 134 | SLC4A4 | SLC4A4 | 21238 | -0.072 | -0.2298 | Yes |
| 135 | NKX2-2 | NKX2-2 | 21263 | -0.073 | -0.2162 | Yes |
| 136 | PLLP | PLLP | 21296 | -0.075 | -0.2026 | Yes |
| 137 | GPR19 | GPR19 | 21330 | -0.077 | -0.1886 | Yes |
| 138 | RAP1GAP | RAP1GAP | 21462 | -0.092 | -0.1763 | Yes |
| 139 | CSPG5 | CSPG5 | 21531 | -0.108 | -0.1579 | Yes |
| 140 | GAS1 | GAS1 | 21567 | -0.130 | -0.1334 | Yes |
| 141 | TTYH1 | TTYH1 | 21573 | -0.137 | -0.1061 | Yes |
| 142 | TIMP4 | TIMP4 | 21580 | -0.147 | -0.0770 | Yes |
| 143 | C1orf61 | C1orf61 | 21592 | -0.181 | -0.0412 | Yes |
| 144 | ASCL1 | ASCL1 | 21596 | -0.206 | 0.0000 | Yes |