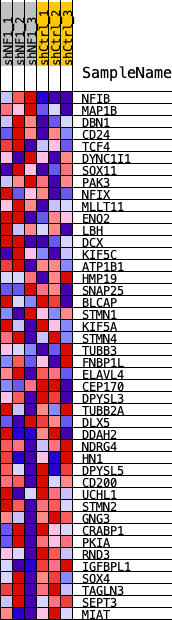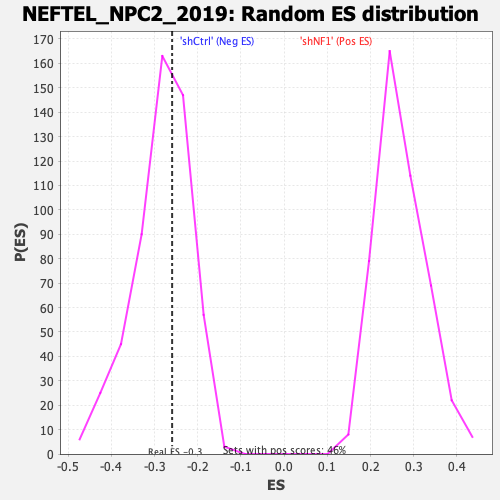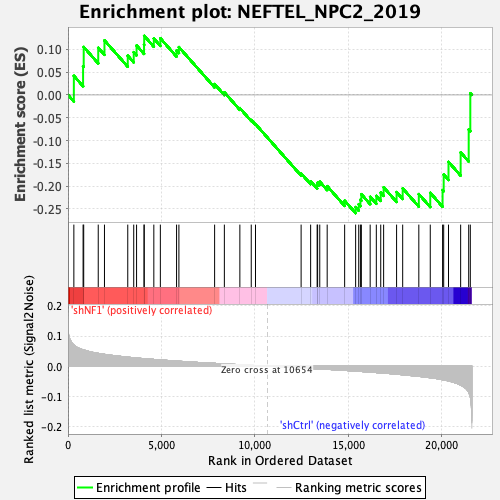
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | NEFTEL_NPC2_2019 |
| Enrichment Score (ES) | -0.25949636 |
| Normalized Enrichment Score (NES) | -0.9178193 |
| Nominal p-value | 0.6044776 |
| FDR q-value | 0.703177 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NFIB | NFIB | 313 | 0.070 | 0.0420 | No |
| 2 | MAP1B | MAP1B | 807 | 0.054 | 0.0632 | No |
| 3 | DBN1 | DBN1 | 840 | 0.054 | 0.1053 | No |
| 4 | CD24 | CD24 | 1617 | 0.042 | 0.1035 | No |
| 5 | TCF4 | TCF4 | 1950 | 0.039 | 0.1198 | No |
| 6 | DYNC1I1 | DYNC1I1 | 3196 | 0.030 | 0.0861 | No |
| 7 | SOX11 | SOX11 | 3515 | 0.028 | 0.0939 | No |
| 8 | PAK3 | PAK3 | 3667 | 0.027 | 0.1086 | No |
| 9 | NFIX | NFIX | 4067 | 0.025 | 0.1101 | No |
| 10 | MLLT11 | MLLT11 | 4075 | 0.025 | 0.1297 | No |
| 11 | ENO2 | ENO2 | 4590 | 0.022 | 0.1238 | No |
| 12 | LBH | LBH | 4939 | 0.021 | 0.1243 | No |
| 13 | DCX | DCX | 5806 | 0.017 | 0.0977 | No |
| 14 | KIF5C | KIF5C | 5931 | 0.016 | 0.1050 | No |
| 15 | ATP1B1 | ATP1B1 | 7841 | 0.009 | 0.0238 | No |
| 16 | HMP19 | HMP19 | 8362 | 0.007 | 0.0057 | No |
| 17 | SNAP25 | SNAP25 | 9191 | 0.005 | -0.0290 | No |
| 18 | BLCAP | BLCAP | 9800 | 0.003 | -0.0550 | No |
| 19 | STMN1 | STMN1 | 10027 | 0.002 | -0.0640 | No |
| 20 | KIF5A | KIF5A | 12467 | -0.006 | -0.1723 | No |
| 21 | STMN4 | STMN4 | 12976 | -0.008 | -0.1897 | No |
| 22 | TUBB3 | TUBB3 | 13324 | -0.009 | -0.1987 | No |
| 23 | FNBP1L | FNBP1L | 13350 | -0.009 | -0.1927 | No |
| 24 | ELAVL4 | ELAVL4 | 13464 | -0.009 | -0.1905 | No |
| 25 | CEP170 | CEP170 | 13861 | -0.011 | -0.2003 | No |
| 26 | DPYSL3 | DPYSL3 | 14795 | -0.014 | -0.2323 | No |
| 27 | TUBB2A | TUBB2A | 15383 | -0.016 | -0.2463 | Yes |
| 28 | DLX5 | DLX5 | 15548 | -0.017 | -0.2400 | Yes |
| 29 | DDAH2 | DDAH2 | 15638 | -0.017 | -0.2300 | Yes |
| 30 | NDRG4 | NDRG4 | 15689 | -0.018 | -0.2180 | Yes |
| 31 | HN1 | HN1 | 16160 | -0.020 | -0.2237 | Yes |
| 32 | DPYSL5 | DPYSL5 | 16489 | -0.021 | -0.2217 | Yes |
| 33 | CD200 | CD200 | 16725 | -0.022 | -0.2144 | Yes |
| 34 | UCHL1 | UCHL1 | 16880 | -0.023 | -0.2028 | Yes |
| 35 | STMN2 | STMN2 | 17573 | -0.027 | -0.2133 | Yes |
| 36 | GNG3 | GNG3 | 17899 | -0.029 | -0.2052 | Yes |
| 37 | CRABP1 | CRABP1 | 18762 | -0.034 | -0.2177 | Yes |
| 38 | PKIA | PKIA | 19374 | -0.038 | -0.2149 | Yes |
| 39 | RND3 | RND3 | 20033 | -0.045 | -0.2090 | Yes |
| 40 | IGFBPL1 | IGFBPL1 | 20092 | -0.046 | -0.1748 | Yes |
| 41 | SOX4 | SOX4 | 20350 | -0.049 | -0.1470 | Yes |
| 42 | TAGLN3 | TAGLN3 | 20999 | -0.063 | -0.1262 | Yes |
| 43 | SEPT3 | SEPT3 | 21430 | -0.087 | -0.0758 | Yes |
| 44 | MIAT | MIAT | 21516 | -0.103 | 0.0037 | Yes |