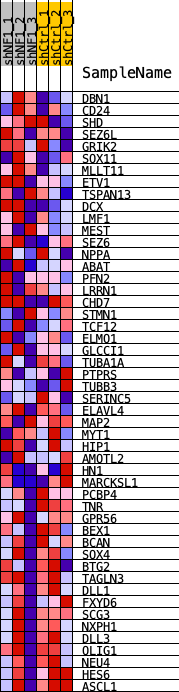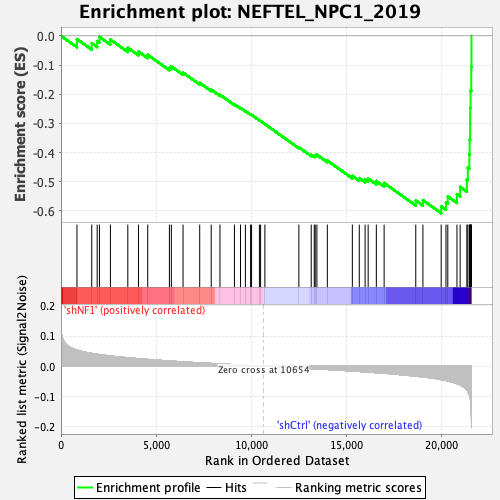
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | NEFTEL_NPC1_2019 |
| Enrichment Score (ES) | -0.60791695 |
| Normalized Enrichment Score (NES) | -2.203602 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | DBN1 | DBN1 | 840 | 0.054 | -0.0111 | No |
| 2 | CD24 | CD24 | 1617 | 0.042 | -0.0251 | No |
| 3 | SHD | SHD | 1903 | 0.039 | -0.0179 | No |
| 4 | SEZ6L | SEZ6L | 2022 | 0.038 | -0.0034 | No |
| 5 | GRIK2 | GRIK2 | 2600 | 0.034 | -0.0127 | No |
| 6 | SOX11 | SOX11 | 3515 | 0.028 | -0.0407 | No |
| 7 | MLLT11 | MLLT11 | 4075 | 0.025 | -0.0539 | No |
| 8 | ETV1 | ETV1 | 4563 | 0.022 | -0.0649 | No |
| 9 | TSPAN13 | TSPAN13 | 5703 | 0.017 | -0.1089 | No |
| 10 | DCX | DCX | 5806 | 0.017 | -0.1049 | No |
| 11 | LMF1 | LMF1 | 6421 | 0.014 | -0.1261 | No |
| 12 | MEST | MEST | 7298 | 0.011 | -0.1611 | No |
| 13 | SEZ6 | SEZ6 | 7903 | 0.009 | -0.1845 | No |
| 14 | NPPA | NPPA | 8364 | 0.007 | -0.2020 | No |
| 15 | ABAT | ABAT | 9125 | 0.005 | -0.2348 | No |
| 16 | PFN2 | PFN2 | 9445 | 0.004 | -0.2476 | No |
| 17 | LRRN1 | LRRN1 | 9701 | 0.003 | -0.2579 | No |
| 18 | CHD7 | CHD7 | 9974 | 0.002 | -0.2694 | No |
| 19 | STMN1 | STMN1 | 10027 | 0.002 | -0.2708 | No |
| 20 | TCF12 | TCF12 | 10438 | 0.001 | -0.2895 | No |
| 21 | ELMO1 | ELMO1 | 10501 | 0.000 | -0.2921 | No |
| 22 | GLCCI1 | GLCCI1 | 10727 | -0.000 | -0.3025 | No |
| 23 | TUBA1A | TUBA1A | 12515 | -0.006 | -0.3822 | No |
| 24 | PTPRS | PTPRS | 13164 | -0.008 | -0.4081 | No |
| 25 | TUBB3 | TUBB3 | 13324 | -0.009 | -0.4109 | No |
| 26 | SERINC5 | SERINC5 | 13375 | -0.009 | -0.4086 | No |
| 27 | ELAVL4 | ELAVL4 | 13464 | -0.009 | -0.4079 | No |
| 28 | MAP2 | MAP2 | 14012 | -0.011 | -0.4275 | No |
| 29 | MYT1 | MYT1 | 15328 | -0.016 | -0.4802 | No |
| 30 | HIP1 | HIP1 | 15693 | -0.018 | -0.4879 | No |
| 31 | AMOTL2 | AMOTL2 | 16000 | -0.019 | -0.4922 | No |
| 32 | HN1 | HN1 | 16160 | -0.020 | -0.4893 | No |
| 33 | MARCKSL1 | MARCKSL1 | 16590 | -0.022 | -0.4979 | No |
| 34 | PCBP4 | PCBP4 | 17000 | -0.024 | -0.5046 | No |
| 35 | TNR | TNR | 18665 | -0.033 | -0.5645 | No |
| 36 | GPR56 | GPR56 | 19041 | -0.036 | -0.5634 | No |
| 37 | BEX1 | BEX1 | 20002 | -0.045 | -0.5847 | Yes |
| 38 | BCAN | BCAN | 20254 | -0.048 | -0.5717 | Yes |
| 39 | SOX4 | SOX4 | 20350 | -0.049 | -0.5506 | Yes |
| 40 | BTG2 | BTG2 | 20832 | -0.058 | -0.5428 | Yes |
| 41 | TAGLN3 | TAGLN3 | 20999 | -0.063 | -0.5179 | Yes |
| 42 | DLL1 | DLL1 | 21352 | -0.079 | -0.4932 | Yes |
| 43 | FXYD6 | FXYD6 | 21404 | -0.084 | -0.4520 | Yes |
| 44 | SCG3 | SCG3 | 21484 | -0.096 | -0.4061 | Yes |
| 45 | NXPH1 | NXPH1 | 21499 | -0.099 | -0.3554 | Yes |
| 46 | DLL3 | DLL3 | 21526 | -0.105 | -0.3021 | Yes |
| 47 | OLIG1 | OLIG1 | 21528 | -0.106 | -0.2472 | Yes |
| 48 | NEU4 | NEU4 | 21549 | -0.115 | -0.1882 | Yes |
| 49 | HES6 | HES6 | 21586 | -0.160 | -0.1066 | Yes |
| 50 | ASCL1 | ASCL1 | 21596 | -0.206 | 0.0000 | Yes |