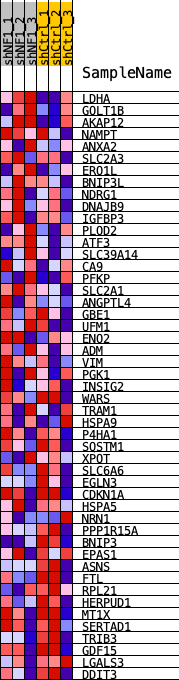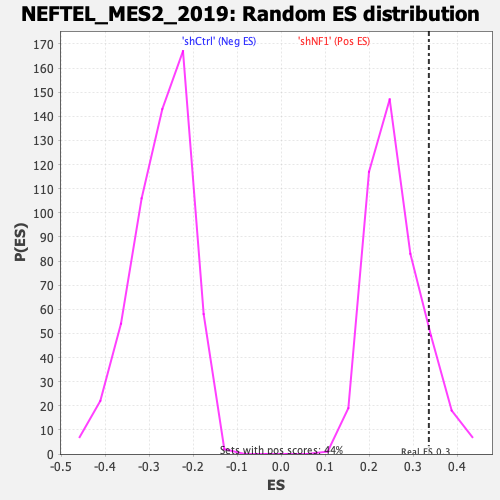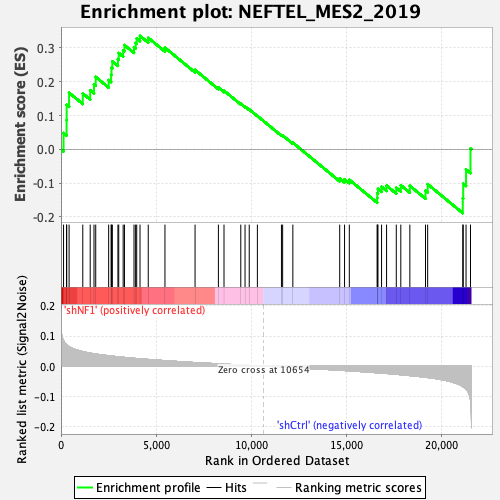
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shNF1 |
| GeneSet | NEFTEL_MES2_2019 |
| Enrichment Score (ES) | 0.3357599 |
| Normalized Enrichment Score (NES) | 1.3002596 |
| Nominal p-value | 0.120181404 |
| FDR q-value | 0.06835011 |
| FWER p-Value | 0.571 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LDHA | LDHA | 134 | 0.084 | 0.0483 | Yes |
| 2 | GOLT1B | GOLT1B | 293 | 0.071 | 0.0865 | Yes |
| 3 | AKAP12 | AKAP12 | 299 | 0.070 | 0.1316 | Yes |
| 4 | NAMPT | NAMPT | 426 | 0.065 | 0.1676 | Yes |
| 5 | ANXA2 | ANXA2 | 1146 | 0.048 | 0.1653 | Yes |
| 6 | SLC2A3 | SLC2A3 | 1536 | 0.043 | 0.1751 | Yes |
| 7 | ERO1L | ERO1L | 1736 | 0.041 | 0.1925 | Yes |
| 8 | BNIP3L | BNIP3L | 1827 | 0.040 | 0.2143 | Yes |
| 9 | NDRG1 | NDRG1 | 2507 | 0.034 | 0.2050 | Yes |
| 10 | DNAJB9 | DNAJB9 | 2636 | 0.033 | 0.2207 | Yes |
| 11 | IGFBP3 | IGFBP3 | 2656 | 0.033 | 0.2413 | Yes |
| 12 | PLOD2 | PLOD2 | 2702 | 0.033 | 0.2605 | Yes |
| 13 | ATF3 | ATF3 | 2995 | 0.031 | 0.2670 | Yes |
| 14 | SLC39A14 | SLC39A14 | 3037 | 0.031 | 0.2850 | Yes |
| 15 | CA9 | CA9 | 3270 | 0.029 | 0.2931 | Yes |
| 16 | PFKP | PFKP | 3339 | 0.029 | 0.3086 | Yes |
| 17 | SLC2A1 | SLC2A1 | 3841 | 0.026 | 0.3021 | Yes |
| 18 | ANGPTL4 | ANGPTL4 | 3926 | 0.025 | 0.3146 | Yes |
| 19 | GBE1 | GBE1 | 3983 | 0.025 | 0.3282 | Yes |
| 20 | UFM1 | UFM1 | 4157 | 0.024 | 0.3358 | Yes |
| 21 | ENO2 | ENO2 | 4590 | 0.022 | 0.3300 | No |
| 22 | ADM | ADM | 5469 | 0.018 | 0.3010 | No |
| 23 | VIM | VIM | 7053 | 0.012 | 0.2351 | No |
| 24 | PGK1 | PGK1 | 8281 | 0.008 | 0.1831 | No |
| 25 | INSIG2 | INSIG2 | 8576 | 0.007 | 0.1737 | No |
| 26 | WARS | WARS | 9459 | 0.004 | 0.1352 | No |
| 27 | TRAM1 | TRAM1 | 9683 | 0.003 | 0.1268 | No |
| 28 | HSPA9 | HSPA9 | 9903 | 0.002 | 0.1181 | No |
| 29 | P4HA1 | P4HA1 | 10332 | 0.001 | 0.0989 | No |
| 30 | SQSTM1 | SQSTM1 | 11598 | -0.003 | 0.0421 | No |
| 31 | XPOT | XPOT | 11661 | -0.003 | 0.0414 | No |
| 32 | SLC6A6 | SLC6A6 | 12198 | -0.005 | 0.0197 | No |
| 33 | EGLN3 | EGLN3 | 14663 | -0.014 | -0.0859 | No |
| 34 | CDKN1A | CDKN1A | 14918 | -0.014 | -0.0883 | No |
| 35 | HSPA5 | HSPA5 | 15169 | -0.015 | -0.0899 | No |
| 36 | NRN1 | NRN1 | 16637 | -0.022 | -0.1438 | No |
| 37 | PPP1R15A | PPP1R15A | 16643 | -0.022 | -0.1298 | No |
| 38 | BNIP3 | BNIP3 | 16676 | -0.022 | -0.1169 | No |
| 39 | EPAS1 | EPAS1 | 16865 | -0.023 | -0.1107 | No |
| 40 | ASNS | ASNS | 17129 | -0.024 | -0.1073 | No |
| 41 | FTL | FTL | 17640 | -0.027 | -0.1135 | No |
| 42 | RPL21 | RPL21 | 17878 | -0.029 | -0.1060 | No |
| 43 | HERPUD1 | HERPUD1 | 18354 | -0.031 | -0.1078 | No |
| 44 | MT1X | MT1X | 19177 | -0.037 | -0.1221 | No |
| 45 | SERTAD1 | SERTAD1 | 19291 | -0.038 | -0.1030 | No |
| 46 | TRIB3 | TRIB3 | 21138 | -0.068 | -0.1449 | No |
| 47 | GDF15 | GDF15 | 21162 | -0.069 | -0.1016 | No |
| 48 | LGALS3 | LGALS3 | 21310 | -0.076 | -0.0595 | No |
| 49 | DDIT3 | DDIT3 | 21544 | -0.113 | 0.0024 | No |