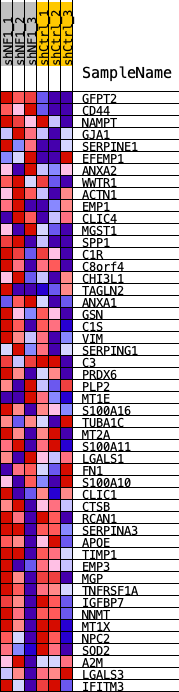
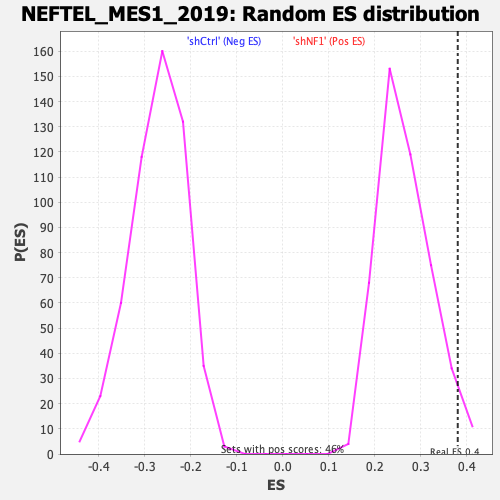
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shNF1 |
| GeneSet | NEFTEL_MES1_2019 |
| Enrichment Score (ES) | 0.38040766 |
| Normalized Enrichment Score (NES) | 1.4326468 |
| Nominal p-value | 0.03448276 |
| FDR q-value | 0.028057724 |
| FWER p-Value | 0.243 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GFPT2 | GFPT2 | 4 | 0.141 | 0.0927 | Yes |
| 2 | CD44 | CD44 | 141 | 0.084 | 0.1415 | Yes |
| 3 | NAMPT | NAMPT | 426 | 0.065 | 0.1710 | Yes |
| 4 | GJA1 | GJA1 | 705 | 0.056 | 0.1951 | Yes |
| 5 | SERPINE1 | SERPINE1 | 725 | 0.056 | 0.2310 | Yes |
| 6 | EFEMP1 | EFEMP1 | 973 | 0.051 | 0.2532 | Yes |
| 7 | ANXA2 | ANXA2 | 1146 | 0.048 | 0.2770 | Yes |
| 8 | WWTR1 | WWTR1 | 1155 | 0.048 | 0.3083 | Yes |
| 9 | ACTN1 | ACTN1 | 1227 | 0.047 | 0.3359 | Yes |
| 10 | EMP1 | EMP1 | 1560 | 0.043 | 0.3487 | Yes |
| 11 | CLIC4 | CLIC4 | 2172 | 0.037 | 0.3448 | Yes |
| 12 | MGST1 | MGST1 | 2364 | 0.036 | 0.3593 | Yes |
| 13 | SPP1 | SPP1 | 2411 | 0.035 | 0.3804 | Yes |
| 14 | C1R | C1R | 4214 | 0.024 | 0.3125 | No |
| 15 | C8orf4 | C8orf4 | 4495 | 0.023 | 0.3144 | No |
| 16 | CHI3L1 | CHI3L1 | 4752 | 0.021 | 0.3166 | No |
| 17 | TAGLN2 | TAGLN2 | 4844 | 0.021 | 0.3262 | No |
| 18 | ANXA1 | ANXA1 | 5293 | 0.019 | 0.3179 | No |
| 19 | GSN | GSN | 6634 | 0.013 | 0.2645 | No |
| 20 | C1S | C1S | 6766 | 0.013 | 0.2669 | No |
| 21 | VIM | VIM | 7053 | 0.012 | 0.2614 | No |
| 22 | SERPING1 | SERPING1 | 7077 | 0.012 | 0.2680 | No |
| 23 | C3 | C3 | 7114 | 0.012 | 0.2740 | No |
| 24 | PRDX6 | PRDX6 | 7145 | 0.011 | 0.2802 | No |
| 25 | PLP2 | PLP2 | 8380 | 0.007 | 0.2277 | No |
| 26 | MT1E | MT1E | 8410 | 0.007 | 0.2311 | No |
| 27 | S100A16 | S100A16 | 9336 | 0.004 | 0.1910 | No |
| 28 | TUBA1C | TUBA1C | 10493 | 0.001 | 0.1376 | No |
| 29 | MT2A | MT2A | 10578 | 0.000 | 0.1339 | No |
| 30 | S100A11 | S100A11 | 10703 | -0.000 | 0.1282 | No |
| 31 | LGALS1 | LGALS1 | 11128 | -0.001 | 0.1095 | No |
| 32 | FN1 | FN1 | 11144 | -0.002 | 0.1098 | No |
| 33 | S100A10 | S100A10 | 11216 | -0.002 | 0.1077 | No |
| 34 | CLIC1 | CLIC1 | 11509 | -0.003 | 0.0959 | No |
| 35 | CTSB | CTSB | 12661 | -0.007 | 0.0468 | No |
| 36 | RCAN1 | RCAN1 | 12732 | -0.007 | 0.0480 | No |
| 37 | SERPINA3 | SERPINA3 | 14162 | -0.012 | -0.0106 | No |
| 38 | APOE | APOE | 15188 | -0.016 | -0.0479 | No |
| 39 | TIMP1 | TIMP1 | 16033 | -0.019 | -0.0745 | No |
| 40 | EMP3 | EMP3 | 16399 | -0.021 | -0.0777 | No |
| 41 | MGP | MGP | 17727 | -0.028 | -0.1211 | No |
| 42 | TNFRSF1A | TNFRSF1A | 18376 | -0.031 | -0.1305 | No |
| 43 | IGFBP7 | IGFBP7 | 18392 | -0.032 | -0.1104 | No |
| 44 | NNMT | NNMT | 18514 | -0.032 | -0.0948 | No |
| 45 | MT1X | MT1X | 19177 | -0.037 | -0.1012 | No |
| 46 | NPC2 | NPC2 | 20205 | -0.047 | -0.1179 | No |
| 47 | SOD2 | SOD2 | 20301 | -0.048 | -0.0905 | No |
| 48 | A2M | A2M | 20660 | -0.055 | -0.0711 | No |
| 49 | LGALS3 | LGALS3 | 21310 | -0.076 | -0.0513 | No |
| 50 | IFITM3 | IFITM3 | 21498 | -0.098 | 0.0045 | No |