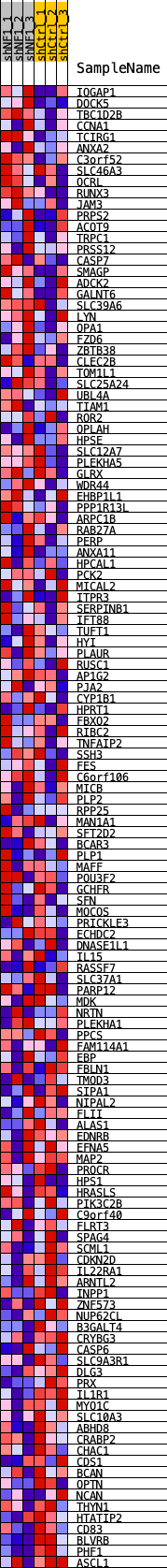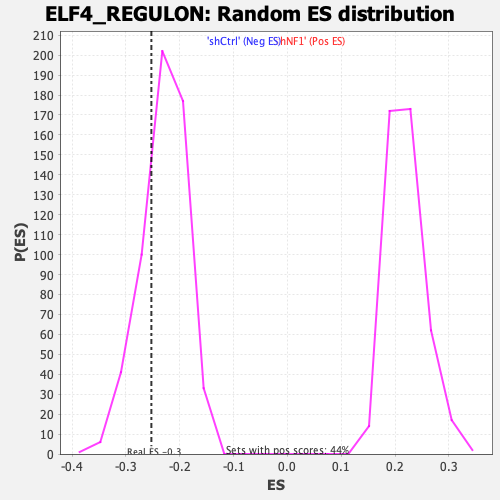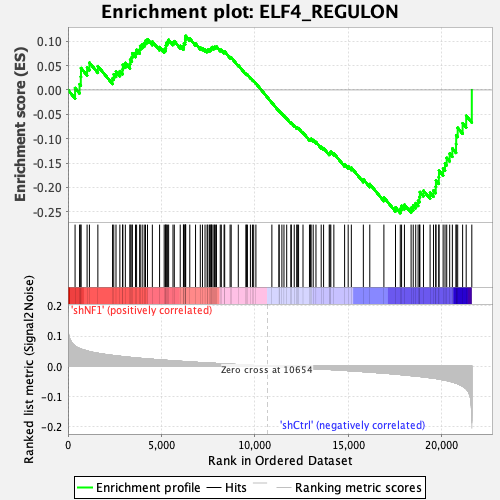
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | ELF4_REGULON |
| Enrichment Score (ES) | -0.2526686 |
| Normalized Enrichment Score (NES) | -1.1024442 |
| Nominal p-value | 0.25357142 |
| FDR q-value | 0.40713987 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IQGAP1 | IQGAP1 | 378 | 0.067 | 0.0040 | No |
| 2 | DOCK5 | DOCK5 | 619 | 0.058 | 0.0117 | No |
| 3 | TBC1D2B | TBC1D2B | 685 | 0.057 | 0.0271 | No |
| 4 | CCNA1 | CCNA1 | 695 | 0.056 | 0.0449 | No |
| 5 | TCIRG1 | TCIRG1 | 1021 | 0.050 | 0.0461 | No |
| 6 | ANXA2 | ANXA2 | 1146 | 0.048 | 0.0560 | No |
| 7 | C3orf52 | C3orf52 | 1597 | 0.042 | 0.0488 | No |
| 8 | SLC46A3 | SLC46A3 | 2383 | 0.035 | 0.0237 | No |
| 9 | OCRL | OCRL | 2447 | 0.035 | 0.0321 | No |
| 10 | RUNX3 | RUNX3 | 2567 | 0.034 | 0.0376 | No |
| 11 | JAM3 | JAM3 | 2774 | 0.033 | 0.0386 | No |
| 12 | PRPS2 | PRPS2 | 2917 | 0.032 | 0.0422 | No |
| 13 | ACOT9 | ACOT9 | 2934 | 0.031 | 0.0516 | No |
| 14 | TRPC1 | TRPC1 | 3066 | 0.031 | 0.0555 | No |
| 15 | PRSS12 | PRSS12 | 3313 | 0.029 | 0.0534 | No |
| 16 | CASP7 | CASP7 | 3325 | 0.029 | 0.0623 | No |
| 17 | SMAGP | SMAGP | 3414 | 0.028 | 0.0673 | No |
| 18 | ADCK2 | ADCK2 | 3441 | 0.028 | 0.0753 | No |
| 19 | GALNT6 | GALNT6 | 3611 | 0.027 | 0.0762 | No |
| 20 | SLC39A6 | SLC39A6 | 3668 | 0.027 | 0.0823 | No |
| 21 | LYN | LYN | 3834 | 0.026 | 0.0830 | No |
| 22 | OPA1 | OPA1 | 3860 | 0.026 | 0.0902 | No |
| 23 | FZD6 | FZD6 | 3968 | 0.025 | 0.0934 | No |
| 24 | ZBTB38 | ZBTB38 | 4076 | 0.025 | 0.0964 | No |
| 25 | CLEC2B | CLEC2B | 4145 | 0.024 | 0.1011 | No |
| 26 | TOM1L1 | TOM1L1 | 4252 | 0.024 | 0.1039 | No |
| 27 | SLC25A24 | SLC25A24 | 4511 | 0.023 | 0.0992 | No |
| 28 | UBL4A | UBL4A | 4898 | 0.021 | 0.0879 | No |
| 29 | TIAM1 | TIAM1 | 5144 | 0.020 | 0.0829 | No |
| 30 | ROR2 | ROR2 | 5221 | 0.019 | 0.0856 | No |
| 31 | OPLAH | OPLAH | 5233 | 0.019 | 0.0913 | No |
| 32 | HPSE | HPSE | 5256 | 0.019 | 0.0965 | No |
| 33 | SLC12A7 | SLC12A7 | 5328 | 0.019 | 0.0993 | No |
| 34 | PLEKHA5 | PLEKHA5 | 5382 | 0.018 | 0.1028 | No |
| 35 | GLRX | GLRX | 5615 | 0.017 | 0.0977 | No |
| 36 | WDR44 | WDR44 | 5685 | 0.017 | 0.1000 | No |
| 37 | EHBP1L1 | EHBP1L1 | 6002 | 0.016 | 0.0904 | No |
| 38 | PPP1R13L | PPP1R13L | 6173 | 0.015 | 0.0874 | No |
| 39 | ARPC1B | ARPC1B | 6192 | 0.015 | 0.0915 | No |
| 40 | RAB27A | RAB27A | 6204 | 0.015 | 0.0959 | No |
| 41 | PERP | PERP | 6265 | 0.015 | 0.0979 | No |
| 42 | ANXA11 | ANXA11 | 6270 | 0.015 | 0.1025 | No |
| 43 | HPCAL1 | HPCAL1 | 6281 | 0.015 | 0.1068 | No |
| 44 | PCK2 | PCK2 | 6295 | 0.015 | 0.1109 | No |
| 45 | MICAL2 | MICAL2 | 6513 | 0.014 | 0.1053 | No |
| 46 | ITPR3 | ITPR3 | 6821 | 0.013 | 0.0951 | No |
| 47 | SERPINB1 | SERPINB1 | 7075 | 0.012 | 0.0871 | No |
| 48 | IFT88 | IFT88 | 7191 | 0.011 | 0.0854 | No |
| 49 | TUFT1 | TUFT1 | 7328 | 0.011 | 0.0826 | No |
| 50 | HYI | HYI | 7439 | 0.010 | 0.0809 | No |
| 51 | PLAUR | PLAUR | 7477 | 0.010 | 0.0825 | No |
| 52 | RUSC1 | RUSC1 | 7563 | 0.010 | 0.0818 | No |
| 53 | AP1G2 | AP1G2 | 7607 | 0.010 | 0.0830 | No |
| 54 | PJA2 | PJA2 | 7649 | 0.010 | 0.0843 | No |
| 55 | CYP1B1 | CYP1B1 | 7673 | 0.010 | 0.0864 | No |
| 56 | HPRT1 | HPRT1 | 7706 | 0.010 | 0.0880 | No |
| 57 | FBXO2 | FBXO2 | 7804 | 0.009 | 0.0865 | No |
| 58 | RIBC2 | RIBC2 | 7830 | 0.009 | 0.0883 | No |
| 59 | TNFAIP2 | TNFAIP2 | 7899 | 0.009 | 0.0880 | No |
| 60 | SSH3 | SSH3 | 7939 | 0.009 | 0.0890 | No |
| 61 | FES | FES | 8143 | 0.008 | 0.0822 | No |
| 62 | C6orf106 | C6orf106 | 8199 | 0.008 | 0.0822 | No |
| 63 | MICB | MICB | 8347 | 0.007 | 0.0778 | No |
| 64 | PLP2 | PLP2 | 8380 | 0.007 | 0.0786 | No |
| 65 | RPP25 | RPP25 | 8669 | 0.006 | 0.0673 | No |
| 66 | MAN1A1 | MAN1A1 | 8727 | 0.006 | 0.0666 | No |
| 67 | SFT2D2 | SFT2D2 | 9107 | 0.005 | 0.0506 | No |
| 68 | BCAR3 | BCAR3 | 9513 | 0.004 | 0.0329 | No |
| 69 | PLP1 | PLP1 | 9543 | 0.004 | 0.0327 | No |
| 70 | MAFF | MAFF | 9596 | 0.003 | 0.0313 | No |
| 71 | POU3F2 | POU3F2 | 9755 | 0.003 | 0.0249 | No |
| 72 | GCHFR | GCHFR | 9864 | 0.002 | 0.0206 | No |
| 73 | SFN | SFN | 9930 | 0.002 | 0.0183 | No |
| 74 | MOCOS | MOCOS | 10049 | 0.002 | 0.0134 | No |
| 75 | PRICKLE3 | PRICKLE3 | 10903 | -0.001 | -0.0261 | No |
| 76 | ECHDC2 | ECHDC2 | 11286 | -0.002 | -0.0432 | No |
| 77 | DNASE1L1 | DNASE1L1 | 11287 | -0.002 | -0.0426 | No |
| 78 | IL15 | IL15 | 11439 | -0.003 | -0.0488 | No |
| 79 | RASSF7 | RASSF7 | 11550 | -0.003 | -0.0530 | No |
| 80 | SLC37A1 | SLC37A1 | 11696 | -0.003 | -0.0586 | No |
| 81 | PARP12 | PARP12 | 11918 | -0.004 | -0.0676 | No |
| 82 | MDK | MDK | 11944 | -0.004 | -0.0674 | No |
| 83 | NRTN | NRTN | 12099 | -0.005 | -0.0731 | No |
| 84 | PLEKHA1 | PLEKHA1 | 12233 | -0.005 | -0.0776 | No |
| 85 | PPCS | PPCS | 12264 | -0.005 | -0.0773 | No |
| 86 | FAM114A1 | FAM114A1 | 12331 | -0.006 | -0.0785 | No |
| 87 | EBP | EBP | 12568 | -0.006 | -0.0875 | No |
| 88 | FBLN1 | FBLN1 | 12914 | -0.007 | -0.1012 | No |
| 89 | TMOD3 | TMOD3 | 12975 | -0.008 | -0.1015 | No |
| 90 | SIPA1 | SIPA1 | 13000 | -0.008 | -0.1001 | No |
| 91 | NIPAL2 | NIPAL2 | 13113 | -0.008 | -0.1027 | No |
| 92 | FLII | FLII | 13261 | -0.009 | -0.1068 | No |
| 93 | ALAS1 | ALAS1 | 13541 | -0.009 | -0.1167 | No |
| 94 | EDNRB | EDNRB | 13669 | -0.010 | -0.1195 | No |
| 95 | EFNA5 | EFNA5 | 13970 | -0.011 | -0.1299 | No |
| 96 | MAP2 | MAP2 | 14012 | -0.011 | -0.1282 | No |
| 97 | PROCR | PROCR | 14056 | -0.011 | -0.1266 | No |
| 98 | HPS1 | HPS1 | 14218 | -0.012 | -0.1302 | No |
| 99 | HRASLS | HRASLS | 14793 | -0.014 | -0.1524 | No |
| 100 | PIK3C2B | PIK3C2B | 14984 | -0.015 | -0.1565 | No |
| 101 | C9orf40 | C9orf40 | 15153 | -0.015 | -0.1593 | No |
| 102 | FLRT3 | FLRT3 | 15795 | -0.018 | -0.1833 | No |
| 103 | SPAG4 | SPAG4 | 16140 | -0.020 | -0.1929 | No |
| 104 | SCML1 | SCML1 | 16894 | -0.023 | -0.2205 | No |
| 105 | CDKN2D | CDKN2D | 17515 | -0.026 | -0.2408 | No |
| 106 | IL22RA1 | IL22RA1 | 17770 | -0.028 | -0.2437 | Yes |
| 107 | ARNTL2 | ARNTL2 | 17840 | -0.028 | -0.2377 | Yes |
| 108 | INPP1 | INPP1 | 17993 | -0.029 | -0.2353 | Yes |
| 109 | ZNF573 | ZNF573 | 18349 | -0.031 | -0.2417 | Yes |
| 110 | NUP62CL | NUP62CL | 18463 | -0.032 | -0.2366 | Yes |
| 111 | B3GALT4 | B3GALT4 | 18596 | -0.033 | -0.2321 | Yes |
| 112 | CRYBG3 | CRYBG3 | 18724 | -0.034 | -0.2271 | Yes |
| 113 | CASP6 | CASP6 | 18796 | -0.034 | -0.2193 | Yes |
| 114 | SLC9A3R1 | SLC9A3R1 | 18820 | -0.034 | -0.2093 | Yes |
| 115 | DLG3 | DLG3 | 19011 | -0.036 | -0.2066 | Yes |
| 116 | PRX | PRX | 19365 | -0.038 | -0.2106 | Yes |
| 117 | IL1R1 | IL1R1 | 19546 | -0.040 | -0.2061 | Yes |
| 118 | MYO1C | MYO1C | 19667 | -0.041 | -0.1984 | Yes |
| 119 | SLC10A3 | SLC10A3 | 19679 | -0.041 | -0.1856 | Yes |
| 120 | ABHD8 | ABHD8 | 19831 | -0.043 | -0.1788 | Yes |
| 121 | CRABP2 | CRABP2 | 19841 | -0.043 | -0.1654 | Yes |
| 122 | CHAC1 | CHAC1 | 20062 | -0.045 | -0.1610 | Yes |
| 123 | CDS1 | CDS1 | 20163 | -0.046 | -0.1506 | Yes |
| 124 | BCAN | BCAN | 20254 | -0.048 | -0.1393 | Yes |
| 125 | OPTN | OPTN | 20412 | -0.050 | -0.1304 | Yes |
| 126 | NCAN | NCAN | 20556 | -0.053 | -0.1200 | Yes |
| 127 | THYN1 | THYN1 | 20753 | -0.056 | -0.1109 | Yes |
| 128 | HTATIP2 | HTATIP2 | 20761 | -0.057 | -0.0928 | Yes |
| 129 | CD83 | CD83 | 20841 | -0.058 | -0.0776 | Yes |
| 130 | BLVRB | BLVRB | 21104 | -0.066 | -0.0683 | Yes |
| 131 | PHF1 | PHF1 | 21295 | -0.075 | -0.0529 | Yes |
| 132 | ASCL1 | ASCL1 | 21596 | -0.206 | 0.0000 | Yes |