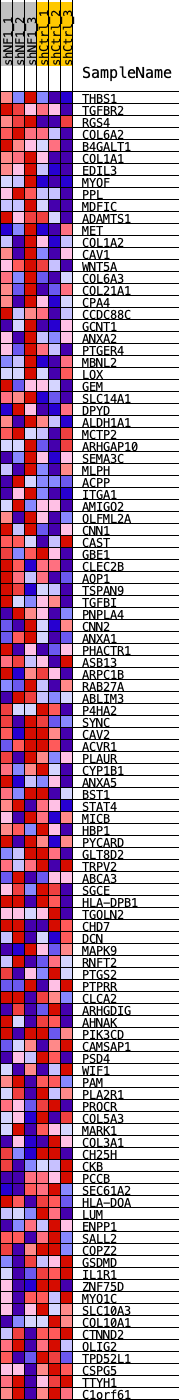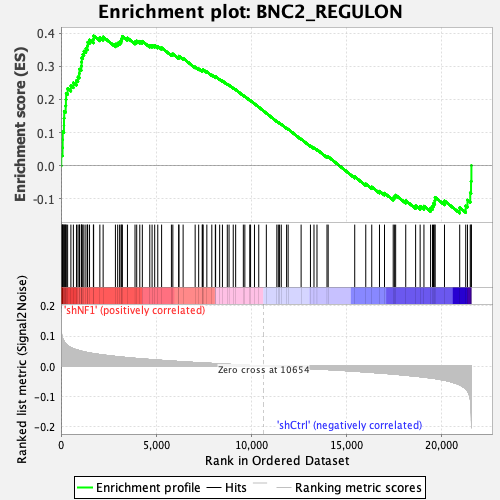
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shNF1 |
| GeneSet | BNC2_REGULON |
| Enrichment Score (ES) | 0.39187753 |
| Normalized Enrichment Score (NES) | 1.7351049 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0015367322 |
| FWER p-Value | 0.009 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | THBS1 | THBS1 | 13 | 0.123 | 0.0323 | Yes |
| 2 | TGFBR2 | TGFBR2 | 80 | 0.093 | 0.0541 | Yes |
| 3 | RGS4 | RGS4 | 88 | 0.092 | 0.0782 | Yes |
| 4 | COL6A2 | COL6A2 | 92 | 0.091 | 0.1023 | Yes |
| 5 | B4GALT1 | B4GALT1 | 155 | 0.083 | 0.1214 | Yes |
| 6 | COL1A1 | COL1A1 | 158 | 0.082 | 0.1432 | Yes |
| 7 | EDIL3 | EDIL3 | 169 | 0.080 | 0.1642 | Yes |
| 8 | MYOF | MYOF | 245 | 0.074 | 0.1806 | Yes |
| 9 | PPL | PPL | 259 | 0.073 | 0.1995 | Yes |
| 10 | MDFIC | MDFIC | 271 | 0.072 | 0.2183 | Yes |
| 11 | ADAMTS1 | ADAMTS1 | 349 | 0.068 | 0.2328 | Yes |
| 12 | MET | MET | 520 | 0.061 | 0.2411 | Yes |
| 13 | COL1A2 | COL1A2 | 651 | 0.057 | 0.2503 | Yes |
| 14 | CAV1 | CAV1 | 814 | 0.054 | 0.2573 | Yes |
| 15 | WNT5A | WNT5A | 892 | 0.053 | 0.2678 | Yes |
| 16 | COL6A3 | COL6A3 | 961 | 0.051 | 0.2783 | Yes |
| 17 | COL21A1 | COL21A1 | 971 | 0.051 | 0.2915 | Yes |
| 18 | CPA4 | CPA4 | 1066 | 0.050 | 0.3003 | Yes |
| 19 | CCDC88C | CCDC88C | 1081 | 0.049 | 0.3128 | Yes |
| 20 | GCNT1 | GCNT1 | 1093 | 0.049 | 0.3254 | Yes |
| 21 | ANXA2 | ANXA2 | 1146 | 0.048 | 0.3359 | Yes |
| 22 | PTGER4 | PTGER4 | 1207 | 0.047 | 0.3457 | Yes |
| 23 | MBNL2 | MBNL2 | 1310 | 0.046 | 0.3531 | Yes |
| 24 | LOX | LOX | 1391 | 0.045 | 0.3613 | Yes |
| 25 | GEM | GEM | 1403 | 0.045 | 0.3727 | Yes |
| 26 | SLC14A1 | SLC14A1 | 1501 | 0.043 | 0.3798 | Yes |
| 27 | DPYD | DPYD | 1704 | 0.041 | 0.3814 | Yes |
| 28 | ALDH1A1 | ALDH1A1 | 1717 | 0.041 | 0.3919 | Yes |
| 29 | MCTP2 | MCTP2 | 2044 | 0.038 | 0.3869 | No |
| 30 | ARHGAP10 | ARHGAP10 | 2217 | 0.037 | 0.3887 | No |
| 31 | SEMA3C | SEMA3C | 2863 | 0.032 | 0.3672 | No |
| 32 | MLPH | MLPH | 2982 | 0.031 | 0.3700 | No |
| 33 | ACPP | ACPP | 3084 | 0.030 | 0.3734 | No |
| 34 | ITGA1 | ITGA1 | 3159 | 0.030 | 0.3780 | No |
| 35 | AMIGO2 | AMIGO2 | 3194 | 0.030 | 0.3843 | No |
| 36 | OLFML2A | OLFML2A | 3228 | 0.030 | 0.3907 | No |
| 37 | CNN1 | CNN1 | 3496 | 0.028 | 0.3857 | No |
| 38 | CAST | CAST | 3894 | 0.026 | 0.3740 | No |
| 39 | GBE1 | GBE1 | 3983 | 0.025 | 0.3766 | No |
| 40 | CLEC2B | CLEC2B | 4145 | 0.024 | 0.3756 | No |
| 41 | AQP1 | AQP1 | 4279 | 0.024 | 0.3757 | No |
| 42 | TSPAN9 | TSPAN9 | 4670 | 0.022 | 0.3634 | No |
| 43 | TGFBI | TGFBI | 4792 | 0.021 | 0.3634 | No |
| 44 | PNPLA4 | PNPLA4 | 4929 | 0.021 | 0.3626 | No |
| 45 | CNN2 | CNN2 | 5091 | 0.020 | 0.3604 | No |
| 46 | ANXA1 | ANXA1 | 5293 | 0.019 | 0.3561 | No |
| 47 | PHACTR1 | PHACTR1 | 5809 | 0.017 | 0.3365 | No |
| 48 | ASB13 | ASB13 | 5884 | 0.016 | 0.3375 | No |
| 49 | ARPC1B | ARPC1B | 6192 | 0.015 | 0.3272 | No |
| 50 | RAB27A | RAB27A | 6204 | 0.015 | 0.3307 | No |
| 51 | ABLIM3 | ABLIM3 | 6425 | 0.014 | 0.3242 | No |
| 52 | P4HA2 | P4HA2 | 7058 | 0.012 | 0.2980 | No |
| 53 | SYNC | SYNC | 7243 | 0.011 | 0.2924 | No |
| 54 | CAV2 | CAV2 | 7429 | 0.010 | 0.2866 | No |
| 55 | ACVR1 | ACVR1 | 7452 | 0.010 | 0.2883 | No |
| 56 | PLAUR | PLAUR | 7477 | 0.010 | 0.2900 | No |
| 57 | CYP1B1 | CYP1B1 | 7673 | 0.010 | 0.2835 | No |
| 58 | ANXA5 | ANXA5 | 7934 | 0.009 | 0.2737 | No |
| 59 | BST1 | BST1 | 8120 | 0.008 | 0.2673 | No |
| 60 | STAT4 | STAT4 | 8135 | 0.008 | 0.2688 | No |
| 61 | MICB | MICB | 8347 | 0.007 | 0.2610 | No |
| 62 | HBP1 | HBP1 | 8493 | 0.007 | 0.2561 | No |
| 63 | PYCARD | PYCARD | 8749 | 0.006 | 0.2458 | No |
| 64 | GLT8D2 | GLT8D2 | 8841 | 0.006 | 0.2432 | No |
| 65 | TRPV2 | TRPV2 | 9062 | 0.005 | 0.2343 | No |
| 66 | ABCA3 | ABCA3 | 9189 | 0.005 | 0.2296 | No |
| 67 | SGCE | SGCE | 9593 | 0.003 | 0.2118 | No |
| 68 | HLA-DPB1 | HLA-DPB1 | 9672 | 0.003 | 0.2090 | No |
| 69 | TGOLN2 | TGOLN2 | 9931 | 0.002 | 0.1975 | No |
| 70 | CHD7 | CHD7 | 9974 | 0.002 | 0.1961 | No |
| 71 | DCN | DCN | 10179 | 0.001 | 0.1870 | No |
| 72 | MAPK9 | MAPK9 | 10404 | 0.001 | 0.1768 | No |
| 73 | RNFT2 | RNFT2 | 10803 | -0.000 | 0.1584 | No |
| 74 | PTGS2 | PTGS2 | 11356 | -0.002 | 0.1333 | No |
| 75 | PTPRR | PTPRR | 11434 | -0.002 | 0.1304 | No |
| 76 | CLCA2 | CLCA2 | 11500 | -0.003 | 0.1281 | No |
| 77 | ARHGDIG | ARHGDIG | 11589 | -0.003 | 0.1248 | No |
| 78 | AHNAK | AHNAK | 11869 | -0.004 | 0.1129 | No |
| 79 | PIK3CD | PIK3CD | 11950 | -0.004 | 0.1103 | No |
| 80 | CAMSAP1 | CAMSAP1 | 12633 | -0.006 | 0.0803 | No |
| 81 | PSD4 | PSD4 | 13124 | -0.008 | 0.0596 | No |
| 82 | WIF1 | WIF1 | 13310 | -0.009 | 0.0533 | No |
| 83 | PAM | PAM | 13468 | -0.009 | 0.0485 | No |
| 84 | PLA2R1 | PLA2R1 | 13994 | -0.011 | 0.0270 | No |
| 85 | PROCR | PROCR | 14056 | -0.011 | 0.0271 | No |
| 86 | COL5A3 | COL5A3 | 15453 | -0.017 | -0.0334 | No |
| 87 | MARK1 | MARK1 | 16034 | -0.019 | -0.0552 | No |
| 88 | COL3A1 | COL3A1 | 16349 | -0.021 | -0.0643 | No |
| 89 | CH25H | CH25H | 16756 | -0.023 | -0.0772 | No |
| 90 | CKB | CKB | 17020 | -0.024 | -0.0831 | No |
| 91 | PCCB | PCCB | 17478 | -0.026 | -0.0974 | No |
| 92 | SEC61A2 | SEC61A2 | 17540 | -0.026 | -0.0932 | No |
| 93 | HLA-DOA | HLA-DOA | 17607 | -0.027 | -0.0891 | No |
| 94 | LUM | LUM | 18137 | -0.030 | -0.1057 | No |
| 95 | ENPP1 | ENPP1 | 18657 | -0.033 | -0.1210 | No |
| 96 | SALL2 | SALL2 | 18895 | -0.035 | -0.1228 | No |
| 97 | COPZ2 | COPZ2 | 19095 | -0.036 | -0.1224 | No |
| 98 | GSDMD | GSDMD | 19445 | -0.039 | -0.1282 | No |
| 99 | IL1R1 | IL1R1 | 19546 | -0.040 | -0.1222 | No |
| 100 | ZNF75D | ZNF75D | 19598 | -0.040 | -0.1139 | No |
| 101 | MYO1C | MYO1C | 19667 | -0.041 | -0.1061 | No |
| 102 | SLC10A3 | SLC10A3 | 19679 | -0.041 | -0.0957 | No |
| 103 | COL10A1 | COL10A1 | 20176 | -0.047 | -0.1063 | No |
| 104 | CTNND2 | CTNND2 | 20977 | -0.062 | -0.1270 | No |
| 105 | OLIG2 | OLIG2 | 21293 | -0.075 | -0.1217 | No |
| 106 | TPD52L1 | TPD52L1 | 21385 | -0.082 | -0.1041 | No |
| 107 | CSPG5 | CSPG5 | 21531 | -0.108 | -0.0820 | No |
| 108 | TTYH1 | TTYH1 | 21573 | -0.137 | -0.0473 | No |
| 109 | C1orf61 | C1orf61 | 21592 | -0.181 | 0.0002 | No |