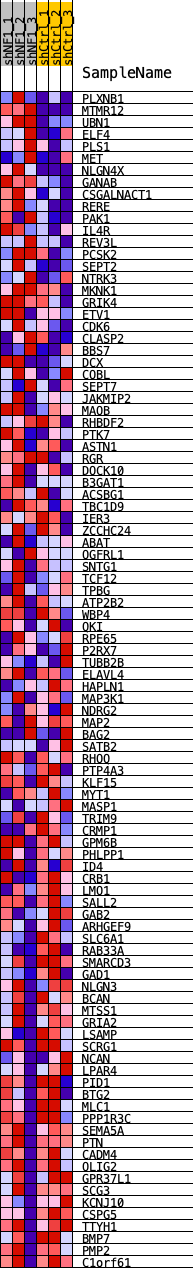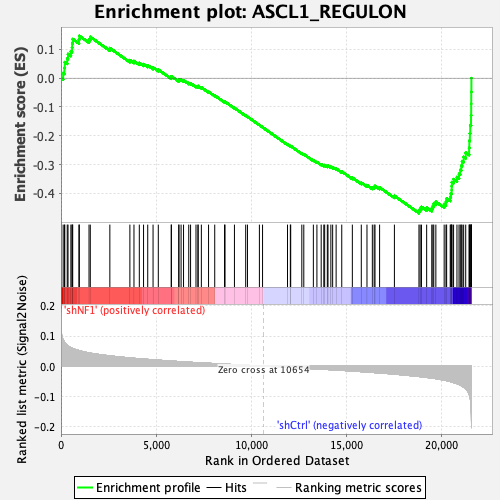
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | shNF1_exprs.shNF1_phenotype.cls #shNF1_versus_shCtrl.shNF1_phenotype.cls #shNF1_versus_shCtrl_repos |
| Phenotype | shNF1_phenotype.cls#shNF1_versus_shCtrl_repos |
| Upregulated in class | shCtrl |
| GeneSet | ASCL1_REGULON |
| Enrichment Score (ES) | -0.4683167 |
| Normalized Enrichment Score (NES) | -1.9404763 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.1666668E-4 |
| FWER p-Value | 0.001 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PLXNB1 | PLXNB1 | 113 | 0.087 | 0.0178 | No |
| 2 | MTMR12 | MTMR12 | 180 | 0.079 | 0.0358 | No |
| 3 | UBN1 | UBN1 | 199 | 0.078 | 0.0556 | No |
| 4 | ELF4 | ELF4 | 328 | 0.069 | 0.0680 | No |
| 5 | PLS1 | PLS1 | 369 | 0.067 | 0.0839 | No |
| 6 | MET | MET | 520 | 0.061 | 0.0930 | No |
| 7 | NLGN4X | NLGN4X | 581 | 0.059 | 0.1060 | No |
| 8 | GANAB | GANAB | 592 | 0.059 | 0.1212 | No |
| 9 | CSGALNACT1 | CSGALNACT1 | 616 | 0.058 | 0.1356 | No |
| 10 | RERE | RERE | 944 | 0.052 | 0.1341 | No |
| 11 | PAK1 | PAK1 | 964 | 0.051 | 0.1468 | No |
| 12 | IL4R | IL4R | 1474 | 0.044 | 0.1348 | No |
| 13 | REV3L | REV3L | 1541 | 0.043 | 0.1431 | No |
| 14 | PCSK2 | PCSK2 | 2570 | 0.034 | 0.1043 | No |
| 15 | SEPT2 | SEPT2 | 3624 | 0.027 | 0.0626 | No |
| 16 | NTRK3 | NTRK3 | 3839 | 0.026 | 0.0595 | No |
| 17 | MKNK1 | MKNK1 | 4120 | 0.024 | 0.0529 | No |
| 18 | GRIK4 | GRIK4 | 4343 | 0.023 | 0.0488 | No |
| 19 | ETV1 | ETV1 | 4563 | 0.022 | 0.0445 | No |
| 20 | CDK6 | CDK6 | 4843 | 0.021 | 0.0371 | No |
| 21 | CLASP2 | CLASP2 | 5119 | 0.020 | 0.0296 | No |
| 22 | BBS7 | BBS7 | 5802 | 0.017 | 0.0023 | No |
| 23 | DCX | DCX | 5806 | 0.017 | 0.0066 | No |
| 24 | COBL | COBL | 6195 | 0.015 | -0.0075 | No |
| 25 | SEPT7 | SEPT7 | 6205 | 0.015 | -0.0039 | No |
| 26 | JAKMIP2 | JAKMIP2 | 6318 | 0.015 | -0.0052 | No |
| 27 | MAOB | MAOB | 6447 | 0.014 | -0.0075 | No |
| 28 | RHBDF2 | RHBDF2 | 6721 | 0.013 | -0.0167 | No |
| 29 | PTK7 | PTK7 | 6817 | 0.013 | -0.0177 | No |
| 30 | ASTN1 | ASTN1 | 7102 | 0.012 | -0.0279 | No |
| 31 | RGR | RGR | 7204 | 0.011 | -0.0296 | No |
| 32 | DOCK10 | DOCK10 | 7217 | 0.011 | -0.0271 | No |
| 33 | B3GAT1 | B3GAT1 | 7384 | 0.011 | -0.0320 | No |
| 34 | ACSBG1 | ACSBG1 | 7763 | 0.009 | -0.0471 | No |
| 35 | TBC1D9 | TBC1D9 | 8088 | 0.008 | -0.0600 | No |
| 36 | IER3 | IER3 | 8602 | 0.007 | -0.0821 | No |
| 37 | ZCCHC24 | ZCCHC24 | 8630 | 0.006 | -0.0817 | No |
| 38 | ABAT | ABAT | 9125 | 0.005 | -0.1034 | No |
| 39 | OGFRL1 | OGFRL1 | 9717 | 0.003 | -0.1301 | No |
| 40 | SNTG1 | SNTG1 | 9810 | 0.003 | -0.1337 | No |
| 41 | TCF12 | TCF12 | 10438 | 0.001 | -0.1626 | No |
| 42 | TPBG | TPBG | 10604 | 0.000 | -0.1703 | No |
| 43 | ATP2B2 | ATP2B2 | 11917 | -0.004 | -0.2302 | No |
| 44 | WBP4 | WBP4 | 12066 | -0.005 | -0.2359 | No |
| 45 | QKI | QKI | 12085 | -0.005 | -0.2355 | No |
| 46 | RPE65 | RPE65 | 12670 | -0.007 | -0.2609 | No |
| 47 | P2RX7 | P2RX7 | 12774 | -0.007 | -0.2638 | No |
| 48 | TUBB2B | TUBB2B | 13277 | -0.009 | -0.2849 | No |
| 49 | ELAVL4 | ELAVL4 | 13464 | -0.009 | -0.2911 | No |
| 50 | HAPLN1 | HAPLN1 | 13689 | -0.010 | -0.2989 | No |
| 51 | MAP3K1 | MAP3K1 | 13827 | -0.010 | -0.3025 | No |
| 52 | NDRG2 | NDRG2 | 13872 | -0.011 | -0.3017 | No |
| 53 | MAP2 | MAP2 | 14012 | -0.011 | -0.3052 | No |
| 54 | BAG2 | BAG2 | 14047 | -0.011 | -0.3038 | No |
| 55 | SATB2 | SATB2 | 14198 | -0.012 | -0.3077 | No |
| 56 | RHOQ | RHOQ | 14292 | -0.012 | -0.3088 | No |
| 57 | PTP4A3 | PTP4A3 | 14475 | -0.013 | -0.3139 | No |
| 58 | KLF15 | KLF15 | 14771 | -0.014 | -0.3239 | No |
| 59 | MYT1 | MYT1 | 15328 | -0.016 | -0.3455 | No |
| 60 | MASP1 | MASP1 | 15799 | -0.018 | -0.3626 | No |
| 61 | TRIM9 | TRIM9 | 16098 | -0.019 | -0.3713 | No |
| 62 | CRMP1 | CRMP1 | 16380 | -0.021 | -0.3788 | No |
| 63 | GPM6B | GPM6B | 16474 | -0.021 | -0.3775 | No |
| 64 | PHLPP1 | PHLPP1 | 16520 | -0.021 | -0.3739 | No |
| 65 | ID4 | ID4 | 16762 | -0.023 | -0.3791 | No |
| 66 | CRB1 | CRB1 | 17541 | -0.026 | -0.4083 | No |
| 67 | LMO1 | LMO1 | 18833 | -0.034 | -0.4592 | Yes |
| 68 | SALL2 | SALL2 | 18895 | -0.035 | -0.4528 | Yes |
| 69 | GAB2 | GAB2 | 18965 | -0.035 | -0.4466 | Yes |
| 70 | ARHGEF9 | ARHGEF9 | 19238 | -0.037 | -0.4493 | Yes |
| 71 | SLC6A1 | SLC6A1 | 19507 | -0.039 | -0.4513 | Yes |
| 72 | RAB33A | RAB33A | 19565 | -0.040 | -0.4434 | Yes |
| 73 | SMARCD3 | SMARCD3 | 19612 | -0.040 | -0.4348 | Yes |
| 74 | GAD1 | GAD1 | 19722 | -0.041 | -0.4289 | Yes |
| 75 | NLGN3 | NLGN3 | 20159 | -0.046 | -0.4368 | Yes |
| 76 | BCAN | BCAN | 20254 | -0.048 | -0.4286 | Yes |
| 77 | MTSS1 | MTSS1 | 20289 | -0.048 | -0.4173 | Yes |
| 78 | GRIA2 | GRIA2 | 20478 | -0.051 | -0.4125 | Yes |
| 79 | LSAMP | LSAMP | 20507 | -0.052 | -0.4001 | Yes |
| 80 | SCRG1 | SCRG1 | 20553 | -0.052 | -0.3883 | Yes |
| 81 | NCAN | NCAN | 20556 | -0.053 | -0.3744 | Yes |
| 82 | LPAR4 | LPAR4 | 20565 | -0.053 | -0.3608 | Yes |
| 83 | PID1 | PID1 | 20654 | -0.054 | -0.3504 | Yes |
| 84 | BTG2 | BTG2 | 20832 | -0.058 | -0.3432 | Yes |
| 85 | MLC1 | MLC1 | 20941 | -0.061 | -0.3321 | Yes |
| 86 | PPP1R3C | PPP1R3C | 21006 | -0.063 | -0.3183 | Yes |
| 87 | SEMA5A | SEMA5A | 21060 | -0.065 | -0.3036 | Yes |
| 88 | PTN | PTN | 21102 | -0.066 | -0.2880 | Yes |
| 89 | CADM4 | CADM4 | 21184 | -0.069 | -0.2733 | Yes |
| 90 | OLIG2 | OLIG2 | 21293 | -0.075 | -0.2585 | Yes |
| 91 | GPR37L1 | GPR37L1 | 21472 | -0.093 | -0.2419 | Yes |
| 92 | SCG3 | SCG3 | 21484 | -0.096 | -0.2171 | Yes |
| 93 | KCNJ10 | KCNJ10 | 21511 | -0.102 | -0.1912 | Yes |
| 94 | CSPG5 | CSPG5 | 21531 | -0.108 | -0.1634 | Yes |
| 95 | TTYH1 | TTYH1 | 21573 | -0.137 | -0.1289 | Yes |
| 96 | BMP7 | BMP7 | 21581 | -0.150 | -0.0893 | Yes |
| 97 | PMP2 | PMP2 | 21584 | -0.157 | -0.0476 | Yes |
| 98 | C1orf61 | C1orf61 | 21592 | -0.181 | 0.0002 | Yes |