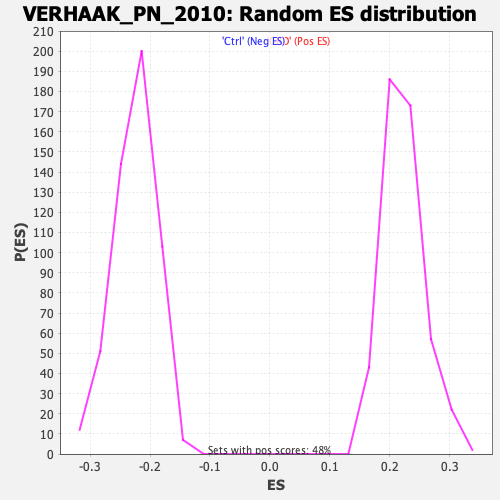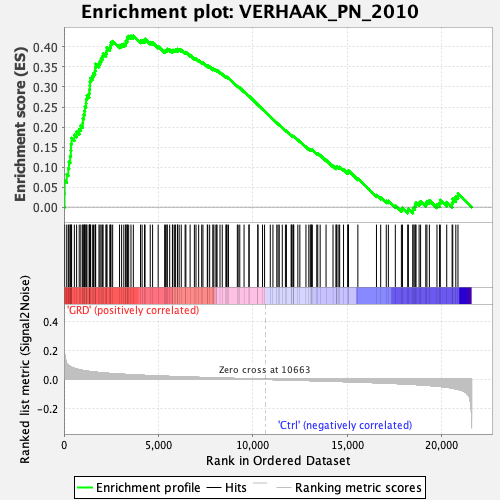
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | GRD |
| GeneSet | VERHAAK_PN_2010 |
| Enrichment Score (ES) | 0.42768008 |
| Normalized Enrichment Score (NES) | 1.9229542 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0011165414 |
| FWER p-Value | 0.002 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GRIA2 | GRIA2 | 27 | 0.178 | 0.0333 | Yes |
| 2 | CLASP2 | CLASP2 | 34 | 0.171 | 0.0661 | Yes |
| 3 | MAP2 | MAP2 | 145 | 0.108 | 0.0820 | Yes |
| 4 | SEZ6L | SEZ6L | 228 | 0.096 | 0.0968 | Yes |
| 5 | TMCC1 | TMCC1 | 264 | 0.093 | 0.1131 | Yes |
| 6 | SRGAP3 | SRGAP3 | 327 | 0.088 | 0.1272 | Yes |
| 7 | MTSS1 | MTSS1 | 362 | 0.085 | 0.1421 | Yes |
| 8 | SNX26 | SNX26 | 373 | 0.084 | 0.1579 | Yes |
| 9 | NCALD | NCALD | 403 | 0.082 | 0.1724 | Yes |
| 10 | NCAM1 | NCAM1 | 549 | 0.075 | 0.1803 | Yes |
| 11 | ZNF286A | ZNF286A | 668 | 0.071 | 0.1884 | Yes |
| 12 | ABAT | ABAT | 806 | 0.066 | 0.1947 | Yes |
| 13 | FXYD6 | FXYD6 | 892 | 0.063 | 0.2030 | Yes |
| 14 | PPM1D | PPM1D | 995 | 0.061 | 0.2100 | Yes |
| 15 | CBX1 | CBX1 | 1000 | 0.060 | 0.2215 | Yes |
| 16 | RALGPS1 | RALGPS1 | 1050 | 0.060 | 0.2307 | Yes |
| 17 | CDKN1B | CDKN1B | 1088 | 0.059 | 0.2404 | Yes |
| 18 | PLCB4 | PLCB4 | 1109 | 0.058 | 0.2508 | Yes |
| 19 | MYB | MYB | 1166 | 0.057 | 0.2593 | Yes |
| 20 | FAM125B | FAM125B | 1173 | 0.057 | 0.2701 | Yes |
| 21 | ZNF184 | ZNF184 | 1217 | 0.056 | 0.2789 | Yes |
| 22 | STMN1 | STMN1 | 1330 | 0.054 | 0.2841 | Yes |
| 23 | SATB1 | SATB1 | 1348 | 0.054 | 0.2937 | Yes |
| 24 | DPYSL4 | DPYSL4 | 1368 | 0.053 | 0.3031 | Yes |
| 25 | NLGN3 | NLGN3 | 1376 | 0.053 | 0.3130 | Yes |
| 26 | TOP2B | TOP2B | 1403 | 0.053 | 0.3220 | Yes |
| 27 | MARCKSL1 | MARCKSL1 | 1500 | 0.051 | 0.3274 | Yes |
| 28 | STMN4 | STMN4 | 1560 | 0.050 | 0.3344 | Yes |
| 29 | TTYH1 | TTYH1 | 1643 | 0.049 | 0.3401 | Yes |
| 30 | MATR3 | MATR3 | 1662 | 0.049 | 0.3488 | Yes |
| 31 | ATP1A3 | ATP1A3 | 1678 | 0.049 | 0.3575 | Yes |
| 32 | DBN1 | DBN1 | 1852 | 0.046 | 0.3584 | Yes |
| 33 | ARHGEF9 | ARHGEF9 | 1905 | 0.046 | 0.3649 | Yes |
| 34 | HN1 | HN1 | 1966 | 0.045 | 0.3708 | Yes |
| 35 | MARCKS | MARCKS | 2032 | 0.044 | 0.3763 | Yes |
| 36 | TAF5 | TAF5 | 2071 | 0.044 | 0.3831 | Yes |
| 37 | SOX10 | SOX10 | 2218 | 0.042 | 0.3844 | Yes |
| 38 | RAB33A | RAB33A | 2265 | 0.042 | 0.3903 | Yes |
| 39 | NRXN2 | NRXN2 | 2272 | 0.042 | 0.3981 | Yes |
| 40 | BCOR | BCOR | 2426 | 0.040 | 0.3987 | Yes |
| 41 | SEC61A2 | SEC61A2 | 2469 | 0.040 | 0.4044 | Yes |
| 42 | SCG3 | SCG3 | 2487 | 0.039 | 0.4112 | Yes |
| 43 | MAST1 | MAST1 | 2578 | 0.039 | 0.4145 | Yes |
| 44 | KLRK1 | KLRK1 | 2943 | 0.036 | 0.4044 | Yes |
| 45 | MYT1 | MYT1 | 3053 | 0.035 | 0.4061 | Yes |
| 46 | C6orf134 | C6orf134 | 3169 | 0.034 | 0.4073 | Yes |
| 47 | C1QL1 | C1QL1 | 3268 | 0.033 | 0.4092 | Yes |
| 48 | FLRT1 | FLRT1 | 3282 | 0.033 | 0.4150 | Yes |
| 49 | LPHN3 | LPHN3 | 3346 | 0.033 | 0.4183 | Yes |
| 50 | TMEFF1 | TMEFF1 | 3356 | 0.033 | 0.4242 | Yes |
| 51 | DUSP26 | DUSP26 | 3426 | 0.032 | 0.4272 | Yes |
| 52 | ALCAM | ALCAM | 3548 | 0.031 | 0.4276 | Yes |
| 53 | WASF1 | WASF1 | 3673 | 0.030 | 0.4277 | Yes |
| 54 | CDK5R1 | CDK5R1 | 4054 | 0.028 | 0.4153 | No |
| 55 | CSPG5 | CSPG5 | 4138 | 0.027 | 0.4168 | No |
| 56 | TOPBP1 | TOPBP1 | 4266 | 0.027 | 0.4160 | No |
| 57 | BCL7A | BCL7A | 4293 | 0.026 | 0.4199 | No |
| 58 | SLCO5A1 | SLCO5A1 | 4568 | 0.025 | 0.4119 | No |
| 59 | PFN2 | PFN2 | 4694 | 0.024 | 0.4108 | No |
| 60 | CRMP1 | CRMP1 | 4989 | 0.023 | 0.4014 | No |
| 61 | C1orf106 | C1orf106 | 5333 | 0.021 | 0.3894 | No |
| 62 | BCAN | BCAN | 5383 | 0.021 | 0.3911 | No |
| 63 | VEZF1 | VEZF1 | 5451 | 0.020 | 0.3919 | No |
| 64 | ACTR1A | ACTR1A | 5470 | 0.020 | 0.3950 | No |
| 65 | PCDH11Y | PCDH11Y | 5599 | 0.020 | 0.3928 | No |
| 66 | PDE10A | PDE10A | 5744 | 0.019 | 0.3898 | No |
| 67 | SOX11 | SOX11 | 5768 | 0.019 | 0.3923 | No |
| 68 | MMP15 | MMP15 | 5862 | 0.018 | 0.3915 | No |
| 69 | RAP2A | RAP2A | 5913 | 0.018 | 0.3927 | No |
| 70 | CHD7 | CHD7 | 6015 | 0.018 | 0.3914 | No |
| 71 | CRB1 | CRB1 | 6017 | 0.018 | 0.3948 | No |
| 72 | ZEB2 | ZEB2 | 6101 | 0.017 | 0.3943 | No |
| 73 | MMP16 | MMP16 | 6212 | 0.017 | 0.3924 | No |
| 74 | DPF1 | DPF1 | 6416 | 0.016 | 0.3860 | No |
| 75 | CSNK1E | CSNK1E | 6457 | 0.016 | 0.3872 | No |
| 76 | PAFAH1B3 | PAFAH1B3 | 6681 | 0.015 | 0.3796 | No |
| 77 | TSPAN3 | TSPAN3 | 6907 | 0.014 | 0.3718 | No |
| 78 | SOX2 | SOX2 | 6993 | 0.013 | 0.3704 | No |
| 79 | CASK | CASK | 7134 | 0.013 | 0.3664 | No |
| 80 | YPEL1 | YPEL1 | 7294 | 0.012 | 0.3614 | No |
| 81 | GPR17 | GPR17 | 7361 | 0.012 | 0.3606 | No |
| 82 | ZNF711 | ZNF711 | 7592 | 0.011 | 0.3520 | No |
| 83 | PELI1 | PELI1 | 7614 | 0.011 | 0.3532 | No |
| 84 | GPM6A | GPM6A | 7717 | 0.011 | 0.3505 | No |
| 85 | EPB41 | EPB41 | 7892 | 0.010 | 0.3444 | No |
| 86 | AMOTL2 | AMOTL2 | 7900 | 0.010 | 0.3460 | No |
| 87 | AOF2 | AOF2 | 8005 | 0.010 | 0.3430 | No |
| 88 | MLLT11 | MLLT11 | 8092 | 0.009 | 0.3408 | No |
| 89 | TTC3 | TTC3 | 8101 | 0.009 | 0.3422 | No |
| 90 | KLRC4 | KLRC4 | 8265 | 0.009 | 0.3362 | No |
| 91 | GNG4 | GNG4 | 8384 | 0.008 | 0.3323 | No |
| 92 | CDC7 | CDC7 | 8577 | 0.008 | 0.3248 | No |
| 93 | P2RX7 | P2RX7 | 8606 | 0.007 | 0.3249 | No |
| 94 | VAX2 | VAX2 | 8641 | 0.007 | 0.3248 | No |
| 95 | SCN3A | SCN3A | 8713 | 0.007 | 0.3228 | No |
| 96 | MYST2 | MYST2 | 9183 | 0.005 | 0.3020 | No |
| 97 | PODXL2 | PODXL2 | 9226 | 0.005 | 0.3010 | No |
| 98 | LRP6 | LRP6 | 9312 | 0.005 | 0.2980 | No |
| 99 | GRID2 | GRID2 | 9539 | 0.004 | 0.2882 | No |
| 100 | GADD45G | GADD45G | 9799 | 0.003 | 0.2767 | No |
| 101 | NOL4 | NOL4 | 9805 | 0.003 | 0.2770 | No |
| 102 | ASCL1 | ASCL1 | 10271 | 0.001 | 0.2556 | No |
| 103 | EPHB1 | EPHB1 | 10273 | 0.001 | 0.2558 | No |
| 104 | ZNF643 | ZNF643 | 10518 | 0.001 | 0.2445 | No |
| 105 | HRASLS | HRASLS | 10626 | 0.000 | 0.2395 | No |
| 106 | BAI3 | BAI3 | 10928 | -0.001 | 0.2256 | No |
| 107 | MAPT | MAPT | 11069 | -0.001 | 0.2194 | No |
| 108 | ATAD5 | ATAD5 | 11274 | -0.002 | 0.2102 | No |
| 109 | HOXD3 | HOXD3 | 11340 | -0.002 | 0.2077 | No |
| 110 | FGF9 | FGF9 | 11403 | -0.003 | 0.2053 | No |
| 111 | ERBB3 | ERBB3 | 11564 | -0.003 | 0.1984 | No |
| 112 | RUFY3 | RUFY3 | 11730 | -0.004 | 0.1915 | No |
| 113 | IL1RAPL1 | IL1RAPL1 | 11783 | -0.004 | 0.1898 | No |
| 114 | SORCS3 | SORCS3 | 12038 | -0.005 | 0.1788 | No |
| 115 | DPP6 | DPP6 | 12045 | -0.005 | 0.1795 | No |
| 116 | TMEM35 | TMEM35 | 12134 | -0.005 | 0.1764 | No |
| 117 | BEX1 | BEX1 | 12138 | -0.005 | 0.1772 | No |
| 118 | C1orf61 | C1orf61 | 12173 | -0.005 | 0.1767 | No |
| 119 | DCX | DCX | 12388 | -0.006 | 0.1678 | No |
| 120 | CXXC4 | CXXC4 | 12495 | -0.006 | 0.1641 | No |
| 121 | PHF16 | PHF16 | 12818 | -0.008 | 0.1506 | No |
| 122 | GABRA3 | GABRA3 | 12970 | -0.008 | 0.1451 | No |
| 123 | RALGPS2 | RALGPS2 | 13046 | -0.008 | 0.1432 | No |
| 124 | PURG | PURG | 13098 | -0.009 | 0.1425 | No |
| 125 | KLRC3 | KLRC3 | 13100 | -0.009 | 0.1441 | No |
| 126 | CLGN | CLGN | 13156 | -0.009 | 0.1433 | No |
| 127 | PCDH11X | PCDH11X | 13392 | -0.010 | 0.1342 | No |
| 128 | SOX4 | SOX4 | 13444 | -0.010 | 0.1337 | No |
| 129 | TOX3 | TOX3 | 13571 | -0.010 | 0.1298 | No |
| 130 | CA10 | CA10 | 13883 | -0.012 | 0.1176 | No |
| 131 | ZNF510 | ZNF510 | 14252 | -0.013 | 0.1029 | No |
| 132 | ZNF248 | ZNF248 | 14416 | -0.014 | 0.0979 | No |
| 133 | HMGB3 | HMGB3 | 14419 | -0.014 | 0.1005 | No |
| 134 | RBPJ | RBPJ | 14462 | -0.014 | 0.1012 | No |
| 135 | RAD21 | RAD21 | 14563 | -0.014 | 0.0993 | No |
| 136 | DLL3 | DLL3 | 14605 | -0.014 | 0.1002 | No |
| 137 | TMEM118 | TMEM118 | 14809 | -0.015 | 0.0936 | No |
| 138 | CKB | CKB | 15032 | -0.016 | 0.0864 | No |
| 139 | HDAC2 | HDAC2 | 15049 | -0.016 | 0.0888 | No |
| 140 | NR0B1 | NR0B1 | 15070 | -0.016 | 0.0911 | No |
| 141 | KIF21B | KIF21B | 15568 | -0.018 | 0.0714 | No |
| 142 | NRXN1 | NRXN1 | 16554 | -0.023 | 0.0299 | No |
| 143 | PPM1E | PPM1E | 16778 | -0.024 | 0.0242 | No |
| 144 | FHOD3 | FHOD3 | 17073 | -0.026 | 0.0155 | No |
| 145 | GSTA4 | GSTA4 | 17184 | -0.026 | 0.0154 | No |
| 146 | REEP1 | REEP1 | 17553 | -0.028 | 0.0037 | No |
| 147 | CAMSAP1L1 | CAMSAP1L1 | 17874 | -0.030 | -0.0054 | No |
| 148 | CNTN1 | CNTN1 | 17931 | -0.031 | -0.0021 | No |
| 149 | ICK | ICK | 18209 | -0.032 | -0.0087 | No |
| 150 | PAK7 | PAK7 | 18244 | -0.033 | -0.0040 | No |
| 151 | MYO10 | MYO10 | 18482 | -0.034 | -0.0084 | No |
| 152 | MCM10 | MCM10 | 18493 | -0.035 | -0.0022 | No |
| 153 | DGKI | DGKI | 18589 | -0.035 | 0.0002 | No |
| 154 | SPTBN2 | SPTBN2 | 18605 | -0.035 | 0.0063 | No |
| 155 | FBXO21 | FBXO21 | 18650 | -0.036 | 0.0111 | No |
| 156 | WDR68 | WDR68 | 18813 | -0.037 | 0.0107 | No |
| 157 | PAK3 | PAK3 | 18892 | -0.038 | 0.0144 | No |
| 158 | DNM3 | DNM3 | 19167 | -0.040 | 0.0094 | No |
| 159 | E2F3 | E2F3 | 19227 | -0.041 | 0.0146 | No |
| 160 | MPPED2 | MPPED2 | 19357 | -0.042 | 0.0167 | No |
| 161 | SLC1A1 | SLC1A1 | 19759 | -0.047 | 0.0070 | No |
| 162 | OLIG2 | OLIG2 | 19896 | -0.049 | 0.0100 | No |
| 163 | CDC25A | CDC25A | 19936 | -0.049 | 0.0177 | No |
| 164 | TNRC4 | TNRC4 | 20276 | -0.054 | 0.0124 | No |
| 165 | LRRTM4 | LRRTM4 | 20562 | -0.060 | 0.0106 | No |
| 166 | ZNF804A | ZNF804A | 20595 | -0.060 | 0.0208 | No |
| 167 | GSK3B | GSK3B | 20757 | -0.065 | 0.0258 | No |
| 168 | FAM110B | FAM110B | 20872 | -0.068 | 0.0337 | No |