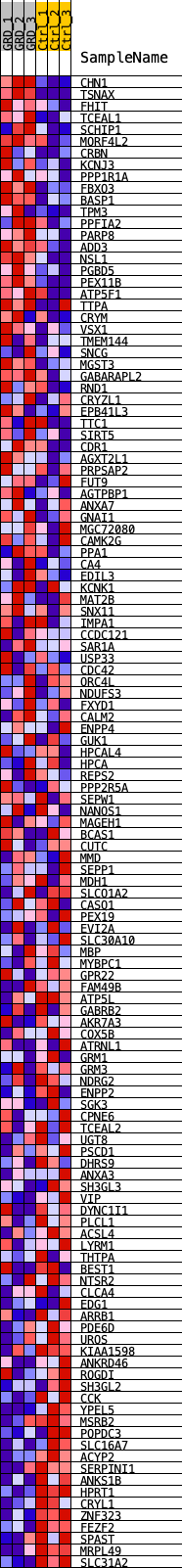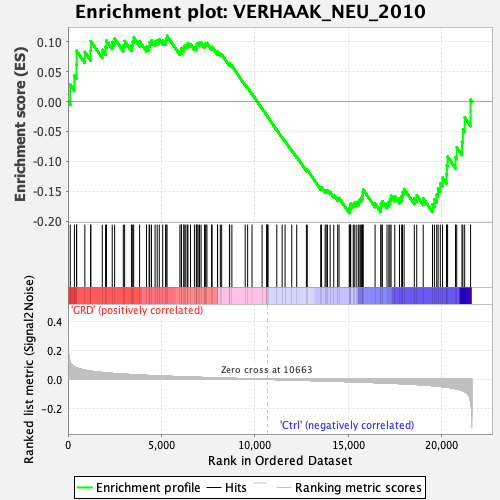
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | VERHAAK_NEU_2010 |
| Enrichment Score (ES) | -0.18673901 |
| Normalized Enrichment Score (NES) | -0.8068953 |
| Nominal p-value | 0.89 |
| FDR q-value | 0.8753801 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CHN1 | CHN1 | 119 | 0.116 | 0.0286 | No |
| 2 | TSNAX | TSNAX | 341 | 0.087 | 0.0437 | No |
| 3 | FHIT | FHIT | 457 | 0.080 | 0.0619 | No |
| 4 | TCEAL1 | TCEAL1 | 463 | 0.080 | 0.0850 | No |
| 5 | SCHIP1 | SCHIP1 | 901 | 0.063 | 0.0831 | No |
| 6 | MORF4L2 | MORF4L2 | 1212 | 0.056 | 0.0851 | No |
| 7 | CRBN | CRBN | 1221 | 0.056 | 0.1012 | No |
| 8 | KCNJ3 | KCNJ3 | 1832 | 0.047 | 0.0865 | No |
| 9 | PPP1R1A | PPP1R1A | 2009 | 0.045 | 0.0914 | No |
| 10 | FBXO3 | FBXO3 | 2057 | 0.044 | 0.1022 | No |
| 11 | BASP1 | BASP1 | 2365 | 0.040 | 0.0998 | No |
| 12 | TPM3 | TPM3 | 2494 | 0.039 | 0.1054 | No |
| 13 | PPFIA2 | PPFIA2 | 2958 | 0.036 | 0.0943 | No |
| 14 | PARP8 | PARP8 | 3029 | 0.035 | 0.1014 | No |
| 15 | ADD3 | ADD3 | 3392 | 0.032 | 0.0940 | No |
| 16 | NSL1 | NSL1 | 3459 | 0.032 | 0.1003 | No |
| 17 | PGBD5 | PGBD5 | 3513 | 0.031 | 0.1070 | No |
| 18 | PEX11B | PEX11B | 3829 | 0.029 | 0.1009 | No |
| 19 | ATP5F1 | ATP5F1 | 4199 | 0.027 | 0.0917 | No |
| 20 | TTPA | TTPA | 4337 | 0.026 | 0.0930 | No |
| 21 | CRYM | CRYM | 4369 | 0.026 | 0.0992 | No |
| 22 | VSX1 | VSX1 | 4464 | 0.025 | 0.1023 | No |
| 23 | TMEM144 | TMEM144 | 4656 | 0.024 | 0.1006 | No |
| 24 | SNCG | SNCG | 4769 | 0.024 | 0.1023 | No |
| 25 | MGST3 | MGST3 | 4881 | 0.023 | 0.1039 | No |
| 26 | GABARAPL2 | GABARAPL2 | 5064 | 0.022 | 0.1019 | No |
| 27 | RND1 | RND1 | 5219 | 0.021 | 0.1011 | No |
| 28 | CRYZL1 | CRYZL1 | 5244 | 0.021 | 0.1062 | No |
| 29 | EPB41L3 | EPB41L3 | 5297 | 0.021 | 0.1100 | No |
| 30 | TTC1 | TTC1 | 5985 | 0.018 | 0.0832 | No |
| 31 | SIRT5 | SIRT5 | 6064 | 0.017 | 0.0847 | No |
| 32 | CDR1 | CDR1 | 6065 | 0.017 | 0.0898 | No |
| 33 | AGXT2L1 | AGXT2L1 | 6196 | 0.017 | 0.0887 | No |
| 34 | PRPSAP2 | PRPSAP2 | 6207 | 0.017 | 0.0932 | No |
| 35 | FUT9 | FUT9 | 6298 | 0.016 | 0.0938 | No |
| 36 | AGTPBP1 | AGTPBP1 | 6379 | 0.016 | 0.0948 | No |
| 37 | ANXA7 | ANXA7 | 6413 | 0.016 | 0.0980 | No |
| 38 | GNAI1 | GNAI1 | 6552 | 0.015 | 0.0961 | No |
| 39 | MGC72080 | MGC72080 | 6759 | 0.014 | 0.0907 | No |
| 40 | CAMK2G | CAMK2G | 6862 | 0.014 | 0.0901 | No |
| 41 | PPA1 | PPA1 | 6887 | 0.014 | 0.0931 | No |
| 42 | CA4 | CA4 | 6891 | 0.014 | 0.0970 | No |
| 43 | EDIL3 | EDIL3 | 6960 | 0.014 | 0.0978 | No |
| 44 | KCNK1 | KCNK1 | 7034 | 0.013 | 0.0983 | No |
| 45 | MAT2B | MAT2B | 7115 | 0.013 | 0.0984 | No |
| 46 | SNX11 | SNX11 | 7303 | 0.012 | 0.0933 | No |
| 47 | IMPA1 | IMPA1 | 7327 | 0.012 | 0.0958 | No |
| 48 | CCDC121 | CCDC121 | 7369 | 0.012 | 0.0974 | No |
| 49 | SAR1A | SAR1A | 7430 | 0.012 | 0.0981 | No |
| 50 | USP33 | USP33 | 7677 | 0.011 | 0.0898 | No |
| 51 | CDC42 | CDC42 | 7706 | 0.011 | 0.0917 | No |
| 52 | ORC4L | ORC4L | 7993 | 0.010 | 0.0812 | No |
| 53 | NDUFS3 | NDUFS3 | 8003 | 0.010 | 0.0836 | No |
| 54 | FXYD1 | FXYD1 | 8146 | 0.009 | 0.0797 | No |
| 55 | CALM2 | CALM2 | 8218 | 0.009 | 0.0789 | No |
| 56 | ENPP4 | ENPP4 | 8634 | 0.007 | 0.0618 | No |
| 57 | GUK1 | GUK1 | 8646 | 0.007 | 0.0634 | No |
| 58 | HPCAL4 | HPCAL4 | 8764 | 0.007 | 0.0599 | No |
| 59 | HPCA | HPCA | 9476 | 0.004 | 0.0281 | No |
| 60 | REPS2 | REPS2 | 9607 | 0.004 | 0.0231 | No |
| 61 | PPP2R5A | PPP2R5A | 9841 | 0.003 | 0.0131 | No |
| 62 | SEPW1 | SEPW1 | 10378 | 0.001 | -0.0116 | No |
| 63 | NANOS1 | NANOS1 | 10631 | 0.000 | -0.0233 | No |
| 64 | MAGEH1 | MAGEH1 | 10633 | 0.000 | -0.0233 | No |
| 65 | BCAS1 | BCAS1 | 10662 | 0.000 | -0.0247 | No |
| 66 | CUTC | CUTC | 10699 | -0.000 | -0.0263 | No |
| 67 | MMD | MMD | 11165 | -0.002 | -0.0474 | No |
| 68 | SEPP1 | SEPP1 | 11451 | -0.003 | -0.0599 | No |
| 69 | MDH1 | MDH1 | 11609 | -0.003 | -0.0662 | No |
| 70 | SLCO1A2 | SLCO1A2 | 11966 | -0.005 | -0.0815 | No |
| 71 | CASQ1 | CASQ1 | 12231 | -0.005 | -0.0922 | No |
| 72 | PEX19 | PEX19 | 12748 | -0.007 | -0.1141 | No |
| 73 | EVI2A | EVI2A | 12796 | -0.007 | -0.1141 | No |
| 74 | SLC30A10 | SLC30A10 | 13514 | -0.010 | -0.1445 | No |
| 75 | MBP | MBP | 13559 | -0.010 | -0.1435 | No |
| 76 | MYBPC1 | MYBPC1 | 13750 | -0.011 | -0.1491 | No |
| 77 | GPR22 | GPR22 | 13807 | -0.011 | -0.1484 | No |
| 78 | FAM49B | FAM49B | 13875 | -0.012 | -0.1482 | No |
| 79 | ATP5L | ATP5L | 14017 | -0.012 | -0.1512 | No |
| 80 | GABRB2 | GABRB2 | 14212 | -0.013 | -0.1564 | No |
| 81 | AKR7A3 | AKR7A3 | 14423 | -0.014 | -0.1622 | No |
| 82 | COX5B | COX5B | 14494 | -0.014 | -0.1614 | No |
| 83 | ATRNL1 | ATRNL1 | 15040 | -0.016 | -0.1820 | Yes |
| 84 | GRM1 | GRM1 | 15067 | -0.016 | -0.1784 | Yes |
| 85 | GRM3 | GRM3 | 15106 | -0.017 | -0.1753 | Yes |
| 86 | NDRG2 | NDRG2 | 15120 | -0.017 | -0.1710 | Yes |
| 87 | ENPP2 | ENPP2 | 15253 | -0.017 | -0.1721 | Yes |
| 88 | SGK3 | SGK3 | 15307 | -0.017 | -0.1695 | Yes |
| 89 | CPNE6 | CPNE6 | 15408 | -0.018 | -0.1690 | Yes |
| 90 | TCEAL2 | TCEAL2 | 15517 | -0.018 | -0.1686 | Yes |
| 91 | UGT8 | UGT8 | 15565 | -0.018 | -0.1654 | Yes |
| 92 | PSCD1 | PSCD1 | 15634 | -0.019 | -0.1631 | Yes |
| 93 | DHRS9 | DHRS9 | 15701 | -0.019 | -0.1605 | Yes |
| 94 | ANXA3 | ANXA3 | 15751 | -0.019 | -0.1571 | Yes |
| 95 | SH3GL3 | SH3GL3 | 15758 | -0.019 | -0.1517 | Yes |
| 96 | VIP | VIP | 15786 | -0.019 | -0.1473 | Yes |
| 97 | DYNC1I1 | DYNC1I1 | 16423 | -0.022 | -0.1703 | Yes |
| 98 | PLCL1 | PLCL1 | 16734 | -0.024 | -0.1777 | Yes |
| 99 | ACSL4 | ACSL4 | 16737 | -0.024 | -0.1708 | Yes |
| 100 | LYRM1 | LYRM1 | 16811 | -0.024 | -0.1670 | Yes |
| 101 | THTPA | THTPA | 17061 | -0.026 | -0.1711 | Yes |
| 102 | BEST1 | BEST1 | 17159 | -0.026 | -0.1679 | Yes |
| 103 | NTSR2 | NTSR2 | 17219 | -0.027 | -0.1629 | Yes |
| 104 | CLCA4 | CLCA4 | 17277 | -0.027 | -0.1577 | Yes |
| 105 | EDG1 | EDG1 | 17474 | -0.028 | -0.1586 | Yes |
| 106 | ARRB1 | ARRB1 | 17728 | -0.029 | -0.1618 | Yes |
| 107 | PDE6D | PDE6D | 17843 | -0.030 | -0.1583 | Yes |
| 108 | UROS | UROS | 17887 | -0.030 | -0.1514 | Yes |
| 109 | KIAA1598 | KIAA1598 | 17974 | -0.031 | -0.1464 | Yes |
| 110 | ANKRD46 | ANKRD46 | 18519 | -0.035 | -0.1615 | Yes |
| 111 | ROGDI | ROGDI | 18649 | -0.036 | -0.1571 | Yes |
| 112 | SH3GL2 | SH3GL2 | 18995 | -0.039 | -0.1618 | Yes |
| 113 | CCK | CCK | 19492 | -0.043 | -0.1721 | Yes |
| 114 | YPEL5 | YPEL5 | 19594 | -0.045 | -0.1637 | Yes |
| 115 | MSRB2 | MSRB2 | 19719 | -0.046 | -0.1560 | Yes |
| 116 | POPDC3 | POPDC3 | 19791 | -0.047 | -0.1455 | Yes |
| 117 | SLC16A7 | SLC16A7 | 19910 | -0.049 | -0.1366 | Yes |
| 118 | ACYP2 | ACYP2 | 20030 | -0.050 | -0.1274 | Yes |
| 119 | SERPINI1 | SERPINI1 | 20241 | -0.053 | -0.1215 | Yes |
| 120 | ANKS1B | ANKS1B | 20262 | -0.054 | -0.1065 | Yes |
| 121 | HPRT1 | HPRT1 | 20301 | -0.055 | -0.0922 | Yes |
| 122 | CRYL1 | CRYL1 | 20724 | -0.064 | -0.0931 | Yes |
| 123 | ZNF323 | ZNF323 | 20788 | -0.066 | -0.0766 | Yes |
| 124 | FEZF2 | FEZF2 | 21069 | -0.076 | -0.0675 | Yes |
| 125 | SPAST | SPAST | 21117 | -0.078 | -0.0468 | Yes |
| 126 | MRPL49 | MRPL49 | 21214 | -0.084 | -0.0265 | Yes |
| 127 | SLC31A2 | SLC31A2 | 21529 | -0.150 | 0.0031 | Yes |