Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | VDR_REGULON |
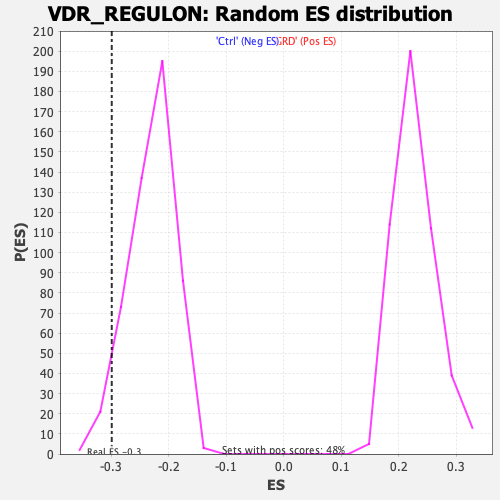
| Enrichment Score (ES) | -0.298967 |
| Normalized Enrichment Score (NES) | -1.2960764 |
| Nominal p-value | 0.044487428 |
| FDR q-value | 0.08163539 |
| FWER p-Value | 0.673 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PTPRD | PTPRD | 9 | 0.241 | 0.0552 | No |
| 2 | HMBOX1 | HMBOX1 | 247 | 0.094 | 0.0660 | No |
| 3 | MPZ | MPZ | 707 | 0.069 | 0.0606 | No |
| 4 | ABAT | ABAT | 806 | 0.066 | 0.0711 | No |
| 5 | ALDH3A1 | ALDH3A1 | 904 | 0.063 | 0.0811 | No |
| 6 | TCF12 | TCF12 | 1195 | 0.057 | 0.0806 | No |
| 7 | PVR | PVR | 1393 | 0.053 | 0.0836 | No |
| 8 | TRIM24 | TRIM24 | 1626 | 0.049 | 0.0843 | No |
| 9 | HLA-DQB1 | HLA-DQB1 | 1749 | 0.048 | 0.0896 | No |
| 10 | GPRC5A | GPRC5A | 2052 | 0.044 | 0.0857 | No |
| 11 | TBX3 | TBX3 | 2301 | 0.041 | 0.0837 | No |
| 12 | MID2 | MID2 | 2308 | 0.041 | 0.0929 | No |
| 13 | FLI1 | FLI1 | 2518 | 0.039 | 0.0922 | No |
| 14 | IPPK | IPPK | 2575 | 0.039 | 0.0985 | No |
| 15 | ABL2 | ABL2 | 2705 | 0.038 | 0.1012 | No |
| 16 | ICOSLG | ICOSLG | 2985 | 0.035 | 0.0964 | No |
| 17 | JUP | JUP | 3350 | 0.033 | 0.0870 | No |
| 18 | TUBB2A | TUBB2A | 3428 | 0.032 | 0.0908 | No |
| 19 | PTPRU | PTPRU | 3570 | 0.031 | 0.0914 | No |
| 20 | STEAP3 | STEAP3 | 3794 | 0.029 | 0.0878 | No |
| 21 | RAD50 | RAD50 | 3942 | 0.028 | 0.0875 | No |
| 22 | ADRB2 | ADRB2 | 4272 | 0.027 | 0.0783 | No |
| 23 | KRT7 | KRT7 | 4491 | 0.025 | 0.0740 | No |
| 24 | LRRC8E | LRRC8E | 4496 | 0.025 | 0.0797 | No |
| 25 | STEAP1 | STEAP1 | 4767 | 0.024 | 0.0725 | No |
| 26 | IL15RA | IL15RA | 4976 | 0.023 | 0.0681 | No |
| 27 | ASPH | ASPH | 5215 | 0.021 | 0.0619 | No |
| 28 | ZFR | ZFR | 5272 | 0.021 | 0.0642 | No |
| 29 | PNMA2 | PNMA2 | 5558 | 0.020 | 0.0555 | No |
| 30 | SOX11 | SOX11 | 5768 | 0.019 | 0.0501 | No |
| 31 | ITPR3 | ITPR3 | 5808 | 0.019 | 0.0526 | No |
| 32 | GALNT6 | GALNT6 | 5829 | 0.019 | 0.0559 | No |
| 33 | S100A8 | S100A8 | 6003 | 0.018 | 0.0520 | No |
| 34 | C9orf3 | C9orf3 | 6008 | 0.018 | 0.0559 | No |
| 35 | CHD7 | CHD7 | 6015 | 0.018 | 0.0597 | No |
| 36 | FAM114A1 | FAM114A1 | 6037 | 0.018 | 0.0627 | No |
| 37 | MARK1 | MARK1 | 6078 | 0.017 | 0.0649 | No |
| 38 | SRGN | SRGN | 6201 | 0.017 | 0.0631 | No |
| 39 | AKR1C1 | AKR1C1 | 6548 | 0.015 | 0.0505 | No |
| 40 | TNFSF9 | TNFSF9 | 6792 | 0.014 | 0.0425 | No |
| 41 | LLGL2 | LLGL2 | 6868 | 0.014 | 0.0422 | No |
| 42 | APOL1 | APOL1 | 6927 | 0.014 | 0.0427 | No |
| 43 | NAV3 | NAV3 | 7054 | 0.013 | 0.0399 | No |
| 44 | CDC42EP1 | CDC42EP1 | 7376 | 0.012 | 0.0277 | No |
| 45 | BCL2A1 | BCL2A1 | 7441 | 0.012 | 0.0274 | No |
| 46 | PHLPP1 | PHLPP1 | 7598 | 0.011 | 0.0227 | No |
| 47 | ME1 | ME1 | 7659 | 0.011 | 0.0224 | No |
| 48 | TGM2 | TGM2 | 7752 | 0.011 | 0.0206 | No |
| 49 | C16orf71 | C16orf71 | 7816 | 0.010 | 0.0201 | No |
| 50 | IL1R1 | IL1R1 | 7833 | 0.010 | 0.0217 | No |
| 51 | IL15 | IL15 | 8040 | 0.009 | 0.0143 | No |
| 52 | CHST15 | CHST15 | 8243 | 0.009 | 0.0069 | No |
| 53 | TRAF3IP1 | TRAF3IP1 | 8394 | 0.008 | 0.0018 | No |
| 54 | LAMC2 | LAMC2 | 8418 | 0.008 | 0.0025 | No |
| 55 | TCIRG1 | TCIRG1 | 8678 | 0.007 | -0.0079 | No |
| 56 | ABLIM3 | ABLIM3 | 8694 | 0.007 | -0.0069 | No |
| 57 | PI15 | PI15 | 8869 | 0.006 | -0.0136 | No |
| 58 | S100A2 | S100A2 | 9027 | 0.006 | -0.0196 | No |
| 59 | TNNC2 | TNNC2 | 9235 | 0.005 | -0.0280 | No |
| 60 | ACSL5 | ACSL5 | 9257 | 0.005 | -0.0278 | No |
| 61 | CDCP1 | CDCP1 | 9398 | 0.005 | -0.0333 | No |
| 62 | TNNT1 | TNNT1 | 9566 | 0.004 | -0.0402 | No |
| 63 | SP140L | SP140L | 9620 | 0.004 | -0.0418 | No |
| 64 | IL24 | IL24 | 9822 | 0.003 | -0.0506 | No |
| 65 | HPCAL1 | HPCAL1 | 10003 | 0.002 | -0.0584 | No |
| 66 | TRIM38 | TRIM38 | 10082 | 0.002 | -0.0616 | No |
| 67 | CBR1 | CBR1 | 10085 | 0.002 | -0.0612 | No |
| 68 | MCOLN3 | MCOLN3 | 10364 | 0.001 | -0.0739 | No |
| 69 | SPAG1 | SPAG1 | 10412 | 0.001 | -0.0759 | No |
| 70 | DCAF7 | DCAF7 | 10450 | 0.001 | -0.0775 | No |
| 71 | CDS1 | CDS1 | 10585 | 0.000 | -0.0836 | No |
| 72 | LAMB3 | LAMB3 | 10602 | 0.000 | -0.0843 | No |
| 73 | AOX1 | AOX1 | 10665 | -0.000 | -0.0872 | No |
| 74 | G0S2 | G0S2 | 11140 | -0.002 | -0.1089 | No |
| 75 | HTR7 | HTR7 | 11232 | -0.002 | -0.1127 | No |
| 76 | KYNU | KYNU | 11298 | -0.002 | -0.1153 | No |
| 77 | MAGEB2 | MAGEB2 | 11801 | -0.004 | -0.1377 | No |
| 78 | GMDS | GMDS | 11876 | -0.004 | -0.1402 | No |
| 79 | HMGA1 | HMGA1 | 11895 | -0.004 | -0.1401 | No |
| 80 | PHF7 | PHF7 | 12048 | -0.005 | -0.1461 | No |
| 81 | TMEM40 | TMEM40 | 12102 | -0.005 | -0.1474 | No |
| 82 | AGPAT2 | AGPAT2 | 12113 | -0.005 | -0.1467 | No |
| 83 | PIK3CD | PIK3CD | 12129 | -0.005 | -0.1462 | No |
| 84 | C1orf61 | C1orf61 | 12173 | -0.005 | -0.1470 | No |
| 85 | TNFAIP2 | TNFAIP2 | 12244 | -0.006 | -0.1490 | No |
| 86 | CXCL3 | CXCL3 | 12327 | -0.006 | -0.1514 | No |
| 87 | PLEK2 | PLEK2 | 12406 | -0.006 | -0.1537 | No |
| 88 | GCNT1 | GCNT1 | 12523 | -0.007 | -0.1576 | No |
| 89 | RAB27A | RAB27A | 12626 | -0.007 | -0.1607 | No |
| 90 | EVI2A | EVI2A | 12796 | -0.007 | -0.1669 | No |
| 91 | GPR39 | GPR39 | 12880 | -0.008 | -0.1690 | No |
| 92 | SERPINA1 | SERPINA1 | 12892 | -0.008 | -0.1677 | No |
| 93 | DOCK2 | DOCK2 | 13017 | -0.008 | -0.1715 | No |
| 94 | ABCA7 | ABCA7 | 13097 | -0.009 | -0.1732 | No |
| 95 | BANK1 | BANK1 | 13225 | -0.009 | -0.1770 | No |
| 96 | DENND3 | DENND3 | 13479 | -0.010 | -0.1865 | No |
| 97 | PKP3 | PKP3 | 13678 | -0.011 | -0.1933 | No |
| 98 | COL17A1 | COL17A1 | 13955 | -0.012 | -0.2034 | No |
| 99 | STARD5 | STARD5 | 14125 | -0.013 | -0.2084 | No |
| 100 | MME | MME | 14310 | -0.013 | -0.2139 | No |
| 101 | SPINT1 | SPINT1 | 14601 | -0.014 | -0.2241 | No |
| 102 | CD55 | CD55 | 15072 | -0.016 | -0.2423 | No |
| 103 | SPAG4 | SPAG4 | 15315 | -0.017 | -0.2495 | No |
| 104 | ID4 | ID4 | 15399 | -0.018 | -0.2493 | No |
| 105 | SERPINB7 | SERPINB7 | 15479 | -0.018 | -0.2488 | No |
| 106 | CCND3 | CCND3 | 15504 | -0.018 | -0.2458 | No |
| 107 | TRAPPC10 | TRAPPC10 | 16153 | -0.021 | -0.2711 | No |
| 108 | SALL2 | SALL2 | 16354 | -0.022 | -0.2753 | No |
| 109 | UBE2Z | UBE2Z | 16451 | -0.023 | -0.2746 | No |
| 110 | FMNL1 | FMNL1 | 16757 | -0.024 | -0.2833 | No |
| 111 | YWHAE | YWHAE | 17050 | -0.026 | -0.2910 | No |
| 112 | PDCD1LG2 | PDCD1LG2 | 17082 | -0.026 | -0.2865 | No |
| 113 | CASP4 | CASP4 | 17111 | -0.026 | -0.2818 | No |
| 114 | STAT6 | STAT6 | 17480 | -0.028 | -0.2925 | Yes |
| 115 | MSX2 | MSX2 | 17495 | -0.028 | -0.2867 | Yes |
| 116 | SVIL | SVIL | 17535 | -0.028 | -0.2820 | Yes |
| 117 | EXPH5 | EXPH5 | 17572 | -0.028 | -0.2771 | Yes |
| 118 | IL2RB | IL2RB | 17658 | -0.029 | -0.2744 | Yes |
| 119 | SYNGR2 | SYNGR2 | 17709 | -0.029 | -0.2700 | Yes |
| 120 | NCAN | NCAN | 17822 | -0.030 | -0.2683 | Yes |
| 121 | RIPK4 | RIPK4 | 17844 | -0.030 | -0.2624 | Yes |
| 122 | TFAP2C | TFAP2C | 17889 | -0.030 | -0.2574 | Yes |
| 123 | PRKCD | PRKCD | 18125 | -0.032 | -0.2611 | Yes |
| 124 | SELPLG | SELPLG | 18133 | -0.032 | -0.2540 | Yes |
| 125 | MAP3K1 | MAP3K1 | 18630 | -0.035 | -0.2690 | Yes |
| 126 | MYO1D | MYO1D | 18698 | -0.036 | -0.2638 | Yes |
| 127 | PRICKLE3 | PRICKLE3 | 18763 | -0.037 | -0.2583 | Yes |
| 128 | SDC1 | SDC1 | 18831 | -0.037 | -0.2528 | Yes |
| 129 | ADARB1 | ADARB1 | 18888 | -0.038 | -0.2468 | Yes |
| 130 | TNFRSF10D | TNFRSF10D | 18900 | -0.038 | -0.2385 | Yes |
| 131 | DNASE1L1 | DNASE1L1 | 18949 | -0.038 | -0.2319 | Yes |
| 132 | LTBR | LTBR | 18956 | -0.038 | -0.2234 | Yes |
| 133 | ENOX2 | ENOX2 | 19508 | -0.044 | -0.2390 | Yes |
| 134 | SLC10A3 | SLC10A3 | 19691 | -0.046 | -0.2369 | Yes |
| 135 | JPH2 | JPH2 | 19706 | -0.046 | -0.2270 | Yes |
| 136 | PLEKHO1 | PLEKHO1 | 19793 | -0.047 | -0.2201 | Yes |
| 137 | TRAF1 | TRAF1 | 19832 | -0.048 | -0.2109 | Yes |
| 138 | C3 | C3 | 20052 | -0.051 | -0.2094 | Yes |
| 139 | CXCL1 | CXCL1 | 20079 | -0.051 | -0.1988 | Yes |
| 140 | VNN1 | VNN1 | 20294 | -0.054 | -0.1962 | Yes |
| 141 | SMARCD2 | SMARCD2 | 20295 | -0.054 | -0.1836 | Yes |
| 142 | PTPRH | PTPRH | 20410 | -0.056 | -0.1759 | Yes |
| 143 | STX11 | STX11 | 20425 | -0.057 | -0.1634 | Yes |
| 144 | IQSEC2 | IQSEC2 | 20471 | -0.058 | -0.1522 | Yes |
| 145 | NFKBIE | NFKBIE | 20540 | -0.059 | -0.1418 | Yes |
| 146 | SLC9A7 | SLC9A7 | 20719 | -0.064 | -0.1353 | Yes |
| 147 | SH3TC1 | SH3TC1 | 20896 | -0.069 | -0.1275 | Yes |
| 148 | IGFBP6 | IGFBP6 | 21022 | -0.074 | -0.1163 | Yes |
| 149 | CPPED1 | CPPED1 | 21119 | -0.078 | -0.1028 | Yes |
| 150 | CYB5R2 | CYB5R2 | 21152 | -0.080 | -0.0858 | Yes |
| 151 | PLP2 | PLP2 | 21203 | -0.083 | -0.0688 | Yes |
| 152 | LTBP2 | LTBP2 | 21266 | -0.088 | -0.0514 | Yes |
| 153 | TPMT | TPMT | 21478 | -0.121 | -0.0332 | Yes |
| 154 | FOSL1 | FOSL1 | 21546 | -0.167 | 0.0023 | Yes |