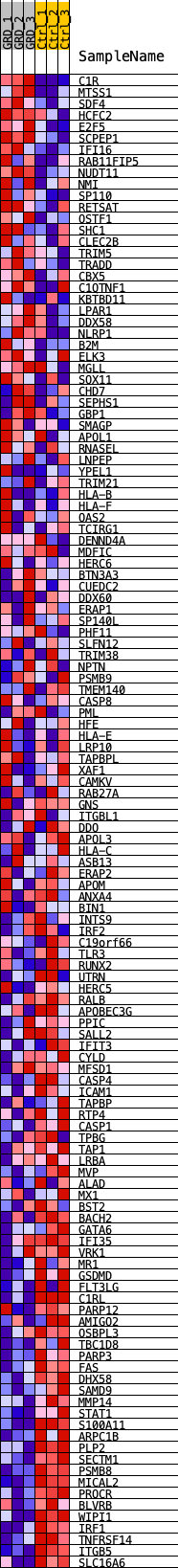
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | SP100_REGULON |
| Enrichment Score (ES) | -0.39346373 |
| Normalized Enrichment Score (NES) | -1.692748 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0040680408 |
| FWER p-Value | 0.028 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | C1R | C1R | 108 | 0.120 | 0.0214 | No |
| 2 | MTSS1 | MTSS1 | 362 | 0.085 | 0.0283 | No |
| 3 | SDF4 | SDF4 | 616 | 0.072 | 0.0323 | No |
| 4 | HCFC2 | HCFC2 | 763 | 0.067 | 0.0403 | No |
| 5 | E2F5 | E2F5 | 815 | 0.065 | 0.0523 | No |
| 6 | SCPEP1 | SCPEP1 | 820 | 0.065 | 0.0664 | No |
| 7 | IFI16 | IFI16 | 831 | 0.065 | 0.0801 | No |
| 8 | RAB11FIP5 | RAB11FIP5 | 875 | 0.064 | 0.0921 | No |
| 9 | NUDT11 | NUDT11 | 1306 | 0.054 | 0.0840 | No |
| 10 | NMI | NMI | 1355 | 0.053 | 0.0935 | No |
| 11 | SP110 | SP110 | 1523 | 0.051 | 0.0969 | No |
| 12 | RETSAT | RETSAT | 1685 | 0.049 | 0.1000 | No |
| 13 | OSTF1 | OSTF1 | 1920 | 0.046 | 0.0992 | No |
| 14 | SHC1 | SHC1 | 2099 | 0.044 | 0.1004 | No |
| 15 | CLEC2B | CLEC2B | 2106 | 0.043 | 0.1097 | No |
| 16 | TRIM5 | TRIM5 | 2928 | 0.036 | 0.0793 | No |
| 17 | TRADD | TRADD | 3580 | 0.031 | 0.0558 | No |
| 18 | CBX5 | CBX5 | 3641 | 0.031 | 0.0597 | No |
| 19 | C1QTNF1 | C1QTNF1 | 3773 | 0.030 | 0.0601 | No |
| 20 | KBTBD11 | KBTBD11 | 3871 | 0.029 | 0.0620 | No |
| 21 | LPAR1 | LPAR1 | 3933 | 0.029 | 0.0654 | No |
| 22 | DDX58 | DDX58 | 3946 | 0.028 | 0.0711 | No |
| 23 | NLRP1 | NLRP1 | 4727 | 0.024 | 0.0400 | No |
| 24 | B2M | B2M | 5076 | 0.022 | 0.0286 | No |
| 25 | ELK3 | ELK3 | 5298 | 0.021 | 0.0229 | No |
| 26 | MGLL | MGLL | 5571 | 0.020 | 0.0146 | No |
| 27 | SOX11 | SOX11 | 5768 | 0.019 | 0.0096 | No |
| 28 | CHD7 | CHD7 | 6015 | 0.018 | 0.0020 | No |
| 29 | SEPHS1 | SEPHS1 | 6256 | 0.017 | -0.0055 | No |
| 30 | GBP1 | GBP1 | 6648 | 0.015 | -0.0205 | No |
| 31 | SMAGP | SMAGP | 6881 | 0.014 | -0.0282 | No |
| 32 | APOL1 | APOL1 | 6927 | 0.014 | -0.0273 | No |
| 33 | RNASEL | RNASEL | 6951 | 0.014 | -0.0254 | No |
| 34 | LNPEP | LNPEP | 7078 | 0.013 | -0.0284 | No |
| 35 | YPEL1 | YPEL1 | 7294 | 0.012 | -0.0357 | No |
| 36 | TRIM21 | TRIM21 | 7539 | 0.011 | -0.0446 | No |
| 37 | HLA-B | HLA-B | 7810 | 0.010 | -0.0548 | No |
| 38 | HLA-F | HLA-F | 8223 | 0.009 | -0.0721 | No |
| 39 | OAS2 | OAS2 | 8425 | 0.008 | -0.0797 | No |
| 40 | TCIRG1 | TCIRG1 | 8678 | 0.007 | -0.0899 | No |
| 41 | DENND4A | DENND4A | 8742 | 0.007 | -0.0913 | No |
| 42 | MDFIC | MDFIC | 8822 | 0.007 | -0.0936 | No |
| 43 | HERC6 | HERC6 | 8899 | 0.006 | -0.0957 | No |
| 44 | BTN3A3 | BTN3A3 | 9289 | 0.005 | -0.1127 | No |
| 45 | CUEDC2 | CUEDC2 | 9311 | 0.005 | -0.1127 | No |
| 46 | DDX60 | DDX60 | 9347 | 0.005 | -0.1132 | No |
| 47 | ERAP1 | ERAP1 | 9374 | 0.005 | -0.1134 | No |
| 48 | SP140L | SP140L | 9620 | 0.004 | -0.1240 | No |
| 49 | PHF11 | PHF11 | 9830 | 0.003 | -0.1331 | No |
| 50 | SLFN12 | SLFN12 | 9836 | 0.003 | -0.1328 | No |
| 51 | TRIM38 | TRIM38 | 10082 | 0.002 | -0.1437 | No |
| 52 | NPTN | NPTN | 10169 | 0.002 | -0.1473 | No |
| 53 | PSMB9 | PSMB9 | 10207 | 0.002 | -0.1487 | No |
| 54 | TMEM140 | TMEM140 | 10443 | 0.001 | -0.1595 | No |
| 55 | CASP8 | CASP8 | 10557 | 0.000 | -0.1647 | No |
| 56 | PML | PML | 10625 | 0.000 | -0.1678 | No |
| 57 | HFE | HFE | 10660 | 0.000 | -0.1693 | No |
| 58 | HLA-E | HLA-E | 11037 | -0.001 | -0.1866 | No |
| 59 | LRP10 | LRP10 | 11474 | -0.003 | -0.2063 | No |
| 60 | TAPBPL | TAPBPL | 11983 | -0.005 | -0.2289 | No |
| 61 | XAF1 | XAF1 | 12073 | -0.005 | -0.2320 | No |
| 62 | CAMKV | CAMKV | 12586 | -0.007 | -0.2544 | No |
| 63 | RAB27A | RAB27A | 12626 | -0.007 | -0.2547 | No |
| 64 | GNS | GNS | 12627 | -0.007 | -0.2532 | No |
| 65 | ITGBL1 | ITGBL1 | 12642 | -0.007 | -0.2523 | No |
| 66 | DDO | DDO | 12952 | -0.008 | -0.2649 | No |
| 67 | APOL3 | APOL3 | 13025 | -0.008 | -0.2665 | No |
| 68 | HLA-C | HLA-C | 13036 | -0.008 | -0.2651 | No |
| 69 | ASB13 | ASB13 | 13194 | -0.009 | -0.2704 | No |
| 70 | ERAP2 | ERAP2 | 13347 | -0.010 | -0.2754 | No |
| 71 | APOM | APOM | 14093 | -0.012 | -0.3074 | No |
| 72 | ANXA4 | ANXA4 | 14597 | -0.014 | -0.3277 | No |
| 73 | BIN1 | BIN1 | 14717 | -0.015 | -0.3299 | No |
| 74 | INTS9 | INTS9 | 15069 | -0.016 | -0.3427 | No |
| 75 | IRF2 | IRF2 | 15223 | -0.017 | -0.3461 | No |
| 76 | C19orf66 | C19orf66 | 15338 | -0.017 | -0.3476 | No |
| 77 | TLR3 | TLR3 | 15426 | -0.018 | -0.3477 | No |
| 78 | RUNX2 | RUNX2 | 15638 | -0.019 | -0.3534 | No |
| 79 | UTRN | UTRN | 15859 | -0.020 | -0.3593 | No |
| 80 | HERC5 | HERC5 | 16012 | -0.021 | -0.3619 | No |
| 81 | RALB | RALB | 16034 | -0.021 | -0.3583 | No |
| 82 | APOBEC3G | APOBEC3G | 16135 | -0.021 | -0.3584 | No |
| 83 | PPIC | PPIC | 16165 | -0.021 | -0.3551 | No |
| 84 | SALL2 | SALL2 | 16354 | -0.022 | -0.3590 | No |
| 85 | IFIT3 | IFIT3 | 16496 | -0.023 | -0.3606 | No |
| 86 | CYLD | CYLD | 16905 | -0.025 | -0.3741 | No |
| 87 | MFSD1 | MFSD1 | 16982 | -0.025 | -0.3721 | No |
| 88 | CASP4 | CASP4 | 17111 | -0.026 | -0.3724 | No |
| 89 | ICAM1 | ICAM1 | 17117 | -0.026 | -0.3669 | No |
| 90 | TAPBP | TAPBP | 17481 | -0.028 | -0.3777 | No |
| 91 | RTP4 | RTP4 | 17640 | -0.029 | -0.3788 | No |
| 92 | CASP1 | CASP1 | 17665 | -0.029 | -0.3735 | No |
| 93 | TPBG | TPBG | 17767 | -0.029 | -0.3718 | No |
| 94 | TAP1 | TAP1 | 18234 | -0.033 | -0.3863 | Yes |
| 95 | LRBA | LRBA | 18305 | -0.033 | -0.3823 | Yes |
| 96 | MVP | MVP | 18318 | -0.033 | -0.3757 | Yes |
| 97 | ALAD | ALAD | 18331 | -0.033 | -0.3689 | Yes |
| 98 | MX1 | MX1 | 18439 | -0.034 | -0.3665 | Yes |
| 99 | BST2 | BST2 | 18775 | -0.037 | -0.3740 | Yes |
| 100 | BACH2 | BACH2 | 18961 | -0.038 | -0.3742 | Yes |
| 101 | GATA6 | GATA6 | 18967 | -0.038 | -0.3660 | Yes |
| 102 | IFI35 | IFI35 | 19250 | -0.041 | -0.3701 | Yes |
| 103 | VRK1 | VRK1 | 19288 | -0.041 | -0.3628 | Yes |
| 104 | MR1 | MR1 | 19310 | -0.042 | -0.3546 | Yes |
| 105 | GSDMD | GSDMD | 19363 | -0.042 | -0.3478 | Yes |
| 106 | FLT3LG | FLT3LG | 19490 | -0.043 | -0.3442 | Yes |
| 107 | C1RL | C1RL | 19665 | -0.046 | -0.3423 | Yes |
| 108 | PARP12 | PARP12 | 19840 | -0.048 | -0.3399 | Yes |
| 109 | AMIGO2 | AMIGO2 | 20174 | -0.052 | -0.3439 | Yes |
| 110 | OSBPL3 | OSBPL3 | 20251 | -0.054 | -0.3357 | Yes |
| 111 | TBC1D8 | TBC1D8 | 20453 | -0.057 | -0.3325 | Yes |
| 112 | PARP3 | PARP3 | 20533 | -0.059 | -0.3233 | Yes |
| 113 | FAS | FAS | 20765 | -0.065 | -0.3197 | Yes |
| 114 | DHX58 | DHX58 | 20776 | -0.065 | -0.3058 | Yes |
| 115 | SAMD9 | SAMD9 | 20789 | -0.066 | -0.2918 | Yes |
| 116 | MMP14 | MMP14 | 20926 | -0.070 | -0.2828 | Yes |
| 117 | STAT1 | STAT1 | 21042 | -0.074 | -0.2718 | Yes |
| 118 | S100A11 | S100A11 | 21047 | -0.075 | -0.2556 | Yes |
| 119 | ARPC1B | ARPC1B | 21177 | -0.082 | -0.2436 | Yes |
| 120 | PLP2 | PLP2 | 21203 | -0.083 | -0.2265 | Yes |
| 121 | SECTM1 | SECTM1 | 21221 | -0.085 | -0.2086 | Yes |
| 122 | PSMB8 | PSMB8 | 21234 | -0.086 | -0.1903 | Yes |
| 123 | MICAL2 | MICAL2 | 21308 | -0.091 | -0.1736 | Yes |
| 124 | PROCR | PROCR | 21321 | -0.093 | -0.1536 | Yes |
| 125 | BLVRB | BLVRB | 21350 | -0.098 | -0.1335 | Yes |
| 126 | WIPI1 | WIPI1 | 21433 | -0.109 | -0.1134 | Yes |
| 127 | IRF1 | IRF1 | 21459 | -0.115 | -0.0894 | Yes |
| 128 | TNFRSF14 | TNFRSF14 | 21471 | -0.118 | -0.0640 | Yes |
| 129 | ITGB5 | ITGB5 | 21506 | -0.140 | -0.0348 | Yes |
| 130 | SLC16A6 | SLC16A6 | 21559 | -0.177 | 0.0017 | Yes |