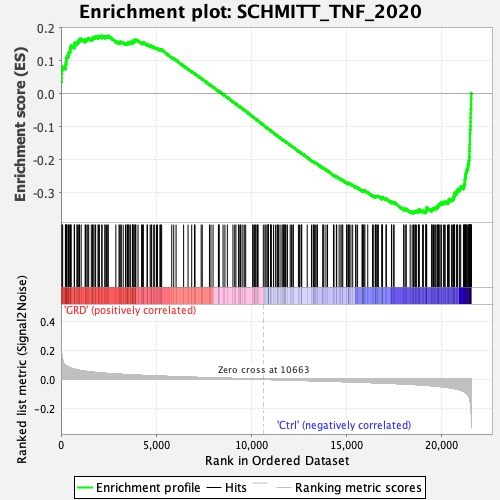
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | SCHMITT_TNF_2020 |
| Enrichment Score (ES) | -0.3609119 |
| Normalized Enrichment Score (NES) | -1.711786 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0039041655 |
| FWER p-Value | 0.022 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SPARCL1 | SPARCL1 | 0 | 0.477 | 0.0365 | No |
| 2 | SLC16A3 | SLC16A3 | 31 | 0.173 | 0.0484 | No |
| 3 | A2M | A2M | 36 | 0.168 | 0.0610 | No |
| 4 | IL1RAP | IL1RAP | 42 | 0.163 | 0.0733 | No |
| 5 | ANXA1 | ANXA1 | 87 | 0.133 | 0.0814 | No |
| 6 | ARRDC4 | ARRDC4 | 232 | 0.096 | 0.0819 | No |
| 7 | FZD5 | FZD5 | 235 | 0.096 | 0.0892 | No |
| 8 | MATN2 | MATN2 | 265 | 0.093 | 0.0949 | No |
| 9 | CD44 | CD44 | 281 | 0.092 | 0.1012 | No |
| 10 | ATCAY | ATCAY | 286 | 0.092 | 0.1080 | No |
| 11 | KLHL5 | KLHL5 | 321 | 0.089 | 0.1132 | No |
| 12 | PRKAR1B | PRKAR1B | 387 | 0.083 | 0.1165 | No |
| 13 | INSR | INSR | 396 | 0.083 | 0.1224 | No |
| 14 | NAMPT | NAMPT | 433 | 0.081 | 0.1269 | No |
| 15 | KLF5 | KLF5 | 486 | 0.078 | 0.1305 | No |
| 16 | PXDN | PXDN | 490 | 0.078 | 0.1363 | No |
| 17 | STAT3 | STAT3 | 496 | 0.078 | 0.1421 | No |
| 18 | SOCS3 | SOCS3 | 538 | 0.076 | 0.1460 | No |
| 19 | SLC16A9 | SLC16A9 | 701 | 0.069 | 0.1437 | No |
| 20 | TNFRSF9 | TNFRSF9 | 706 | 0.069 | 0.1488 | No |
| 21 | MPZ | MPZ | 707 | 0.069 | 0.1540 | No |
| 22 | F3 | F3 | 837 | 0.065 | 0.1529 | No |
| 23 | CSRP2 | CSRP2 | 861 | 0.064 | 0.1568 | No |
| 24 | CDKAL1 | CDKAL1 | 930 | 0.062 | 0.1583 | No |
| 25 | C4A | C4A | 968 | 0.061 | 0.1613 | No |
| 26 | SLC2A3 | SLC2A3 | 977 | 0.061 | 0.1656 | No |
| 27 | CNTNAP1 | CNTNAP1 | 1061 | 0.059 | 0.1662 | No |
| 28 | PODXL | PODXL | 1273 | 0.055 | 0.1605 | No |
| 29 | PTP4A3 | PTP4A3 | 1283 | 0.055 | 0.1643 | No |
| 30 | BIRC2 | BIRC2 | 1360 | 0.053 | 0.1648 | No |
| 31 | RELL1 | RELL1 | 1411 | 0.053 | 0.1664 | No |
| 32 | OSMR | OSMR | 1457 | 0.052 | 0.1683 | No |
| 33 | LRP2 | LRP2 | 1595 | 0.050 | 0.1657 | No |
| 34 | JUNB | JUNB | 1660 | 0.049 | 0.1664 | No |
| 35 | ZNF347 | ZNF347 | 1670 | 0.049 | 0.1697 | No |
| 36 | DACT1 | DACT1 | 1729 | 0.048 | 0.1707 | No |
| 37 | SOD2 | SOD2 | 1797 | 0.047 | 0.1711 | No |
| 38 | ARHGEF16 | ARHGEF16 | 1823 | 0.047 | 0.1735 | No |
| 39 | BCL6 | BCL6 | 1952 | 0.045 | 0.1710 | No |
| 40 | EFTUD2 | EFTUD2 | 1964 | 0.045 | 0.1739 | No |
| 41 | ZIK1 | ZIK1 | 2021 | 0.044 | 0.1747 | No |
| 42 | SEMA4B | SEMA4B | 2149 | 0.043 | 0.1720 | No |
| 43 | ZNF813 | ZNF813 | 2157 | 0.043 | 0.1750 | No |
| 44 | BCL3 | BCL3 | 2307 | 0.041 | 0.1711 | No |
| 45 | IFNGR1 | IFNGR1 | 2319 | 0.041 | 0.1737 | No |
| 46 | RGS16 | RGS16 | 2391 | 0.040 | 0.1735 | No |
| 47 | RPGR | RPGR | 2459 | 0.040 | 0.1733 | No |
| 48 | NCOA7 | NCOA7 | 2485 | 0.039 | 0.1752 | No |
| 49 | CRABP2 | CRABP2 | 2885 | 0.036 | 0.1592 | No |
| 50 | CLIC2 | CLIC2 | 3052 | 0.035 | 0.1541 | No |
| 51 | NUAK2 | NUAK2 | 3103 | 0.034 | 0.1544 | No |
| 52 | PTX3 | PTX3 | 3107 | 0.034 | 0.1569 | No |
| 53 | PDGFRL | PDGFRL | 3174 | 0.034 | 0.1564 | No |
| 54 | PEPD | PEPD | 3273 | 0.033 | 0.1543 | No |
| 55 | SFRP1 | SFRP1 | 3395 | 0.032 | 0.1511 | No |
| 56 | ADAMTS4 | ADAMTS4 | 3421 | 0.032 | 0.1524 | No |
| 57 | GTSF1 | GTSF1 | 3504 | 0.031 | 0.1509 | No |
| 58 | PGBD5 | PGBD5 | 3513 | 0.031 | 0.1529 | No |
| 59 | SNX10 | SNX10 | 3515 | 0.031 | 0.1553 | No |
| 60 | GRB10 | GRB10 | 3563 | 0.031 | 0.1555 | No |
| 61 | ADAMTS1 | ADAMTS1 | 3635 | 0.031 | 0.1545 | No |
| 62 | PMP22 | PMP22 | 3640 | 0.031 | 0.1566 | No |
| 63 | EBI3 | EBI3 | 3755 | 0.030 | 0.1535 | No |
| 64 | BATF3 | BATF3 | 3760 | 0.030 | 0.1556 | No |
| 65 | ZNF229 | ZNF229 | 3771 | 0.030 | 0.1574 | No |
| 66 | S100A10 | S100A10 | 3782 | 0.030 | 0.1592 | No |
| 67 | STAP2 | STAP2 | 3798 | 0.029 | 0.1607 | No |
| 68 | SYNGR3 | SYNGR3 | 3890 | 0.029 | 0.1587 | No |
| 69 | BACE2 | BACE2 | 3897 | 0.029 | 0.1606 | No |
| 70 | NAB2 | NAB2 | 3904 | 0.029 | 0.1625 | No |
| 71 | CDH4 | CDH4 | 3939 | 0.029 | 0.1631 | No |
| 72 | PLAT | PLAT | 4053 | 0.028 | 0.1599 | No |
| 73 | CREB3 | CREB3 | 4238 | 0.027 | 0.1533 | No |
| 74 | ZNF329 | ZNF329 | 4295 | 0.026 | 0.1527 | No |
| 75 | CD83 | CD83 | 4319 | 0.026 | 0.1536 | No |
| 76 | VAX1 | VAX1 | 4322 | 0.026 | 0.1556 | No |
| 77 | APOL6 | APOL6 | 4528 | 0.025 | 0.1479 | No |
| 78 | PRR16 | PRR16 | 4546 | 0.025 | 0.1490 | No |
| 79 | HAS2 | HAS2 | 4692 | 0.024 | 0.1440 | No |
| 80 | ZFP36 | ZFP36 | 4748 | 0.024 | 0.1432 | No |
| 81 | ELL2 | ELL2 | 4749 | 0.024 | 0.1451 | No |
| 82 | SGPP2 | SGPP2 | 4856 | 0.023 | 0.1419 | No |
| 83 | LIFR | LIFR | 4928 | 0.023 | 0.1403 | No |
| 84 | FJX1 | FJX1 | 5024 | 0.022 | 0.1375 | No |
| 85 | B2M | B2M | 5076 | 0.022 | 0.1368 | No |
| 86 | OPTN | OPTN | 5205 | 0.022 | 0.1324 | No |
| 87 | PSMD5 | PSMD5 | 5228 | 0.021 | 0.1330 | No |
| 88 | BMP5 | BMP5 | 5262 | 0.021 | 0.1331 | No |
| 89 | THY1 | THY1 | 5296 | 0.021 | 0.1332 | No |
| 90 | STOM | STOM | 5826 | 0.019 | 0.1097 | No |
| 91 | IL6ST | IL6ST | 5931 | 0.018 | 0.1062 | No |
| 92 | NPNT | NPNT | 6054 | 0.017 | 0.1018 | No |
| 93 | NR4A3 | NR4A3 | 6451 | 0.016 | 0.0845 | No |
| 94 | CSF1 | CSF1 | 6684 | 0.015 | 0.0747 | No |
| 95 | ZNF267 | ZNF267 | 6876 | 0.014 | 0.0668 | No |
| 96 | FZD8 | FZD8 | 7021 | 0.013 | 0.0610 | No |
| 97 | PCDH1 | PCDH1 | 7051 | 0.013 | 0.0607 | No |
| 98 | RBM3 | RBM3 | 7375 | 0.012 | 0.0464 | No |
| 99 | GABRA5 | GABRA5 | 7431 | 0.012 | 0.0448 | No |
| 100 | HLA-B | HLA-B | 7810 | 0.010 | 0.0278 | No |
| 101 | CYP1A1 | CYP1A1 | 7889 | 0.010 | 0.0249 | No |
| 102 | CDC42EP5 | CDC42EP5 | 7999 | 0.010 | 0.0205 | No |
| 103 | VWA1 | VWA1 | 8282 | 0.009 | 0.0079 | No |
| 104 | IL32 | IL32 | 8306 | 0.008 | 0.0075 | No |
| 105 | ZNF611 | ZNF611 | 8319 | 0.008 | 0.0076 | No |
| 106 | EMILIN2 | EMILIN2 | 8526 | 0.008 | -0.0015 | No |
| 107 | CDKN2B | CDKN2B | 8624 | 0.007 | -0.0055 | No |
| 108 | PARP9 | PARP9 | 8752 | 0.007 | -0.0109 | No |
| 109 | PDGFA | PDGFA | 9046 | 0.006 | -0.0243 | No |
| 110 | TSC22D4 | TSC22D4 | 9139 | 0.006 | -0.0282 | No |
| 111 | ETV6 | ETV6 | 9202 | 0.005 | -0.0307 | No |
| 112 | WIPF3 | WIPF3 | 9336 | 0.005 | -0.0365 | No |
| 113 | ERAP1 | ERAP1 | 9374 | 0.005 | -0.0379 | No |
| 114 | TMEM173 | TMEM173 | 9450 | 0.004 | -0.0411 | No |
| 115 | CYP4V2 | CYP4V2 | 9462 | 0.004 | -0.0413 | No |
| 116 | ORAI1 | ORAI1 | 9567 | 0.004 | -0.0459 | No |
| 117 | MITD1 | MITD1 | 9643 | 0.004 | -0.0492 | No |
| 118 | GCLM | GCLM | 9716 | 0.003 | -0.0523 | No |
| 119 | SERPINA3 | SERPINA3 | 10080 | 0.002 | -0.0692 | No |
| 120 | EGFLAM | EGFLAM | 10147 | 0.002 | -0.0721 | No |
| 121 | IER2 | IER2 | 10164 | 0.002 | -0.0727 | No |
| 122 | ZNF28 | ZNF28 | 10202 | 0.002 | -0.0743 | No |
| 123 | NFE2L3 | NFE2L3 | 10242 | 0.001 | -0.0761 | No |
| 124 | C1orf226 | C1orf226 | 10309 | 0.001 | -0.0791 | No |
| 125 | KCNN4 | KCNN4 | 10334 | 0.001 | -0.0801 | No |
| 126 | ITGAV | ITGAV | 10370 | 0.001 | -0.0817 | No |
| 127 | NUDT4 | NUDT4 | 10643 | 0.000 | -0.0945 | No |
| 128 | NFKBIZ | NFKBIZ | 10728 | -0.000 | -0.0984 | No |
| 129 | MGAT4B | MGAT4B | 10778 | -0.000 | -0.1007 | No |
| 130 | ASB9 | ASB9 | 10841 | -0.001 | -0.1035 | No |
| 131 | SPRED3 | SPRED3 | 10899 | -0.001 | -0.1062 | No |
| 132 | LHX9 | LHX9 | 10908 | -0.001 | -0.1065 | No |
| 133 | SORBS1 | SORBS1 | 10911 | -0.001 | -0.1065 | No |
| 134 | OSCP1 | OSCP1 | 10914 | -0.001 | -0.1065 | No |
| 135 | ZNF468 | ZNF468 | 11024 | -0.001 | -0.1116 | No |
| 136 | HLA-E | HLA-E | 11037 | -0.001 | -0.1120 | No |
| 137 | ACSL6 | ACSL6 | 11065 | -0.001 | -0.1132 | No |
| 138 | FOSL2 | FOSL2 | 11178 | -0.002 | -0.1183 | No |
| 139 | NFKB1 | NFKB1 | 11278 | -0.002 | -0.1228 | No |
| 140 | LPGAT1 | LPGAT1 | 11369 | -0.002 | -0.1268 | No |
| 141 | TNFRSF1A | TNFRSF1A | 11380 | -0.003 | -0.1271 | No |
| 142 | CARS | CARS | 11438 | -0.003 | -0.1296 | No |
| 143 | NRG1 | NRG1 | 11523 | -0.003 | -0.1333 | No |
| 144 | NR4A1 | NR4A1 | 11625 | -0.003 | -0.1378 | No |
| 145 | ARHGEF4 | ARHGEF4 | 11675 | -0.004 | -0.1398 | No |
| 146 | ANO9 | ANO9 | 11713 | -0.004 | -0.1413 | No |
| 147 | RAB31 | RAB31 | 11737 | -0.004 | -0.1421 | No |
| 148 | NTN1 | NTN1 | 11776 | -0.004 | -0.1436 | No |
| 149 | LRRC4B | LRRC4B | 11821 | -0.004 | -0.1453 | No |
| 150 | NDUFB8 | NDUFB8 | 11831 | -0.004 | -0.1454 | No |
| 151 | L1CAM | L1CAM | 11912 | -0.004 | -0.1489 | No |
| 152 | ALPL | ALPL | 12058 | -0.005 | -0.1553 | No |
| 153 | PIK3CD | PIK3CD | 12129 | -0.005 | -0.1582 | No |
| 154 | KLHL6 | KLHL6 | 12204 | -0.005 | -0.1613 | No |
| 155 | TRIM8 | TRIM8 | 12222 | -0.005 | -0.1616 | No |
| 156 | LGALS1 | LGALS1 | 12490 | -0.006 | -0.1737 | No |
| 157 | SGK1 | SGK1 | 12551 | -0.007 | -0.1760 | No |
| 158 | ZNF766 | ZNF766 | 12637 | -0.007 | -0.1795 | No |
| 159 | WWTR1 | WWTR1 | 12641 | -0.007 | -0.1791 | No |
| 160 | ZDHHC14 | ZDHHC14 | 12666 | -0.007 | -0.1797 | No |
| 161 | DOC2B | DOC2B | 12953 | -0.008 | -0.1925 | No |
| 162 | FGF2 | FGF2 | 13179 | -0.009 | -0.2024 | No |
| 163 | ZNF528 | ZNF528 | 13265 | -0.009 | -0.2057 | No |
| 164 | IFIH1 | IFIH1 | 13292 | -0.009 | -0.2062 | No |
| 165 | NFKB2 | NFKB2 | 13329 | -0.009 | -0.2071 | No |
| 166 | HIVEP1 | HIVEP1 | 13411 | -0.010 | -0.2102 | No |
| 167 | IRF3 | IRF3 | 13414 | -0.010 | -0.2095 | No |
| 168 | COL9A1 | COL9A1 | 13493 | -0.010 | -0.2124 | No |
| 169 | GAP43 | GAP43 | 13780 | -0.011 | -0.2250 | No |
| 170 | IL4I1 | IL4I1 | 13781 | -0.011 | -0.2242 | No |
| 171 | KLF10 | KLF10 | 13829 | -0.011 | -0.2255 | No |
| 172 | DUSP6 | DUSP6 | 13939 | -0.012 | -0.2297 | No |
| 173 | RELB | RELB | 14022 | -0.012 | -0.2326 | No |
| 174 | GABRB3 | GABRB3 | 14334 | -0.013 | -0.2462 | No |
| 175 | LIMD2 | LIMD2 | 14364 | -0.013 | -0.2466 | No |
| 176 | EMILIN1 | EMILIN1 | 14486 | -0.014 | -0.2512 | No |
| 177 | CITED4 | CITED4 | 14506 | -0.014 | -0.2510 | No |
| 178 | TAF13 | TAF13 | 14634 | -0.014 | -0.2559 | No |
| 179 | LOXL2 | LOXL2 | 14726 | -0.015 | -0.2590 | No |
| 180 | RHPN2 | RHPN2 | 14785 | -0.015 | -0.2606 | No |
| 181 | FAM43A | FAM43A | 14824 | -0.015 | -0.2612 | No |
| 182 | IFITM1 | IFITM1 | 15019 | -0.016 | -0.2691 | No |
| 183 | FADS3 | FADS3 | 15104 | -0.017 | -0.2717 | No |
| 184 | ZNF667 | ZNF667 | 15111 | -0.017 | -0.2707 | No |
| 185 | PLA2G4C | PLA2G4C | 15113 | -0.017 | -0.2695 | No |
| 186 | RGS3 | RGS3 | 15170 | -0.017 | -0.2709 | No |
| 187 | ENPP2 | ENPP2 | 15253 | -0.017 | -0.2734 | No |
| 188 | SMOC2 | SMOC2 | 15346 | -0.017 | -0.2764 | No |
| 189 | VDR | VDR | 15499 | -0.018 | -0.2822 | No |
| 190 | FAM174B | FAM174B | 15521 | -0.018 | -0.2818 | No |
| 191 | CA8 | CA8 | 15603 | -0.019 | -0.2841 | No |
| 192 | AHR | AHR | 15846 | -0.020 | -0.2940 | No |
| 193 | DPP4 | DPP4 | 15849 | -0.020 | -0.2926 | No |
| 194 | ALPK1 | ALPK1 | 15888 | -0.020 | -0.2928 | No |
| 195 | MYO5B | MYO5B | 15898 | -0.020 | -0.2917 | No |
| 196 | MAOB | MAOB | 15939 | -0.020 | -0.2920 | No |
| 197 | CTSS | CTSS | 15988 | -0.020 | -0.2927 | No |
| 198 | NRCAM | NRCAM | 16138 | -0.021 | -0.2981 | No |
| 199 | CAMK2D | CAMK2D | 16390 | -0.022 | -0.3082 | No |
| 200 | ETNK2 | ETNK2 | 16452 | -0.023 | -0.3093 | No |
| 201 | TNIP2 | TNIP2 | 16539 | -0.023 | -0.3116 | No |
| 202 | KIAA0040 | KIAA0040 | 16549 | -0.023 | -0.3103 | No |
| 203 | TNC | TNC | 16569 | -0.023 | -0.3094 | No |
| 204 | GFRA1 | GFRA1 | 16618 | -0.023 | -0.3099 | No |
| 205 | ADAMTS14 | ADAMTS14 | 16661 | -0.024 | -0.3101 | No |
| 206 | ARL6IP5 | ARL6IP5 | 16696 | -0.024 | -0.3099 | No |
| 207 | MGAT3 | MGAT3 | 16883 | -0.025 | -0.3167 | No |
| 208 | OSBPL5 | OSBPL5 | 16892 | -0.025 | -0.3152 | No |
| 209 | CYLD | CYLD | 16905 | -0.025 | -0.3139 | No |
| 210 | ZSWIM4 | ZSWIM4 | 16916 | -0.025 | -0.3124 | No |
| 211 | ROBO1 | ROBO1 | 17097 | -0.026 | -0.3189 | No |
| 212 | ICAM1 | ICAM1 | 17117 | -0.026 | -0.3178 | No |
| 213 | PGM2L1 | PGM2L1 | 17386 | -0.027 | -0.3283 | No |
| 214 | PNKD | PNKD | 17390 | -0.027 | -0.3263 | No |
| 215 | TAPBP | TAPBP | 17481 | -0.028 | -0.3284 | No |
| 216 | PERP | PERP | 17526 | -0.028 | -0.3283 | No |
| 217 | RIPK1 | RIPK1 | 18029 | -0.031 | -0.3495 | No |
| 218 | ABR | ABR | 18044 | -0.031 | -0.3478 | No |
| 219 | SYT12 | SYT12 | 18143 | -0.032 | -0.3500 | No |
| 220 | RASEF | RASEF | 18160 | -0.032 | -0.3483 | No |
| 221 | PTPRE | PTPRE | 18369 | -0.033 | -0.3555 | No |
| 222 | PLEKHA4 | PLEKHA4 | 18474 | -0.034 | -0.3578 | No |
| 223 | C10orf90 | C10orf90 | 18542 | -0.035 | -0.3582 | Yes |
| 224 | MICALL2 | MICALL2 | 18568 | -0.035 | -0.3567 | Yes |
| 225 | TGFB1 | TGFB1 | 18625 | -0.035 | -0.3567 | Yes |
| 226 | PLLP | PLLP | 18644 | -0.036 | -0.3548 | Yes |
| 227 | NFKBIA | NFKBIA | 18697 | -0.036 | -0.3545 | Yes |
| 228 | ADAM19 | ADAM19 | 18801 | -0.037 | -0.3565 | Yes |
| 229 | RNF145 | RNF145 | 18808 | -0.037 | -0.3539 | Yes |
| 230 | GRWD1 | GRWD1 | 18842 | -0.037 | -0.3526 | Yes |
| 231 | BMP2 | BMP2 | 18843 | -0.037 | -0.3498 | Yes |
| 232 | DTX3L | DTX3L | 19022 | -0.039 | -0.3552 | Yes |
| 233 | SEMA3B | SEMA3B | 19064 | -0.039 | -0.3541 | Yes |
| 234 | SYNPO | SYNPO | 19198 | -0.041 | -0.3573 | Yes |
| 235 | APBA1 | APBA1 | 19206 | -0.041 | -0.3545 | Yes |
| 236 | TIFA | TIFA | 19209 | -0.041 | -0.3515 | Yes |
| 237 | IL27RA | IL27RA | 19221 | -0.041 | -0.3489 | Yes |
| 238 | TBC1D10A | TBC1D10A | 19226 | -0.041 | -0.3459 | Yes |
| 239 | RAB35 | RAB35 | 19240 | -0.041 | -0.3434 | Yes |
| 240 | C1orf198 | C1orf198 | 19498 | -0.043 | -0.3521 | Yes |
| 241 | ZNF701 | ZNF701 | 19514 | -0.044 | -0.3495 | Yes |
| 242 | TAGLN2 | TAGLN2 | 19565 | -0.044 | -0.3485 | Yes |
| 243 | CXCL10 | CXCL10 | 19596 | -0.045 | -0.3465 | Yes |
| 244 | KREMEN2 | KREMEN2 | 19663 | -0.045 | -0.3461 | Yes |
| 245 | TMEM151B | TMEM151B | 19693 | -0.046 | -0.3439 | Yes |
| 246 | UGCG | UGCG | 19783 | -0.047 | -0.3445 | Yes |
| 247 | EMP3 | EMP3 | 19792 | -0.047 | -0.3413 | Yes |
| 248 | TRAF1 | TRAF1 | 19832 | -0.048 | -0.3395 | Yes |
| 249 | ZBED1 | ZBED1 | 19862 | -0.048 | -0.3372 | Yes |
| 250 | FAM131B | FAM131B | 19876 | -0.048 | -0.3341 | Yes |
| 251 | TMEM158 | TMEM158 | 19929 | -0.049 | -0.3328 | Yes |
| 252 | IGFBP3 | IGFBP3 | 19998 | -0.050 | -0.3322 | Yes |
| 253 | CLDN4 | CLDN4 | 20006 | -0.050 | -0.3287 | Yes |
| 254 | CITED1 | CITED1 | 20119 | -0.051 | -0.3300 | Yes |
| 255 | LARP6 | LARP6 | 20168 | -0.052 | -0.3283 | Yes |
| 256 | MAP3K11 | MAP3K11 | 20212 | -0.053 | -0.3263 | Yes |
| 257 | CABLES1 | CABLES1 | 20336 | -0.055 | -0.3278 | Yes |
| 258 | PLCD3 | PLCD3 | 20398 | -0.056 | -0.3264 | Yes |
| 259 | IFNGR2 | IFNGR2 | 20399 | -0.056 | -0.3221 | Yes |
| 260 | CD63 | CD63 | 20414 | -0.056 | -0.3184 | Yes |
| 261 | NFKBIE | NFKBIE | 20540 | -0.059 | -0.3198 | Yes |
| 262 | PLEKHF1 | PLEKHF1 | 20606 | -0.061 | -0.3182 | Yes |
| 263 | TRPM8 | TRPM8 | 20649 | -0.062 | -0.3154 | Yes |
| 264 | SQSTM1 | SQSTM1 | 20653 | -0.062 | -0.3108 | Yes |
| 265 | LAPTM5 | LAPTM5 | 20687 | -0.063 | -0.3076 | Yes |
| 266 | RRAS | RRAS | 20702 | -0.064 | -0.3034 | Yes |
| 267 | AGT | AGT | 20709 | -0.064 | -0.2988 | Yes |
| 268 | FAM129A | FAM129A | 20817 | -0.067 | -0.2987 | Yes |
| 269 | RAB13 | RAB13 | 20830 | -0.067 | -0.2941 | Yes |
| 270 | ARNTL2 | ARNTL2 | 20855 | -0.068 | -0.2900 | Yes |
| 271 | ADAMTS9 | ADAMTS9 | 20954 | -0.071 | -0.2892 | Yes |
| 272 | CDKN1A | CDKN1A | 20989 | -0.072 | -0.2853 | Yes |
| 273 | TIMP1 | TIMP1 | 21029 | -0.074 | -0.2814 | Yes |
| 274 | SERTAD1 | SERTAD1 | 21172 | -0.081 | -0.2819 | Yes |
| 275 | CAPN5 | CAPN5 | 21192 | -0.083 | -0.2765 | Yes |
| 276 | DRAM1 | DRAM1 | 21232 | -0.086 | -0.2717 | Yes |
| 277 | ASPHD1 | ASPHD1 | 21242 | -0.086 | -0.2656 | Yes |
| 278 | IRAK2 | IRAK2 | 21246 | -0.087 | -0.2591 | Yes |
| 279 | NBL1 | NBL1 | 21269 | -0.088 | -0.2534 | Yes |
| 280 | PRICKLE1 | PRICKLE1 | 21280 | -0.089 | -0.2471 | Yes |
| 281 | CEBPB | CEBPB | 21289 | -0.090 | -0.2406 | Yes |
| 282 | SH2B3 | SH2B3 | 21319 | -0.093 | -0.2348 | Yes |
| 283 | EGR2 | EGR2 | 21344 | -0.097 | -0.2286 | Yes |
| 284 | LIF | LIF | 21404 | -0.105 | -0.2233 | Yes |
| 285 | LRRN3 | LRRN3 | 21411 | -0.106 | -0.2155 | Yes |
| 286 | WARS | WARS | 21449 | -0.112 | -0.2087 | Yes |
| 287 | IRF1 | IRF1 | 21459 | -0.115 | -0.2003 | Yes |
| 288 | IER3 | IER3 | 21480 | -0.122 | -0.1919 | Yes |
| 289 | SPP1 | SPP1 | 21488 | -0.124 | -0.1827 | Yes |
| 290 | SDC4 | SDC4 | 21489 | -0.125 | -0.1732 | Yes |
| 291 | TNFAIP3 | TNFAIP3 | 21499 | -0.132 | -0.1636 | Yes |
| 292 | PLAU | PLAU | 21507 | -0.141 | -0.1531 | Yes |
| 293 | HMOX1 | HMOX1 | 21512 | -0.144 | -0.1423 | Yes |
| 294 | EMP1 | EMP1 | 21515 | -0.145 | -0.1313 | Yes |
| 295 | SPSB1 | SPSB1 | 21516 | -0.145 | -0.1202 | Yes |
| 296 | AHNAK2 | AHNAK2 | 21523 | -0.148 | -0.1092 | Yes |
| 297 | TNFRSF12A | TNFRSF12A | 21534 | -0.154 | -0.0979 | Yes |
| 298 | ISG20 | ISG20 | 21547 | -0.169 | -0.0855 | Yes |
| 299 | RNF19A | RNF19A | 21550 | -0.172 | -0.0725 | Yes |
| 300 | SERPINE1 | SERPINE1 | 21554 | -0.173 | -0.0594 | Yes |
| 301 | VGF | VGF | 21563 | -0.182 | -0.0458 | Yes |
| 302 | CHI3L1 | CHI3L1 | 21572 | -0.194 | -0.0314 | Yes |
| 303 | RDH10 | RDH10 | 21576 | -0.204 | -0.0159 | Yes |
| 304 | DUSP5 | DUSP5 | 21581 | -0.219 | 0.0007 | Yes |

