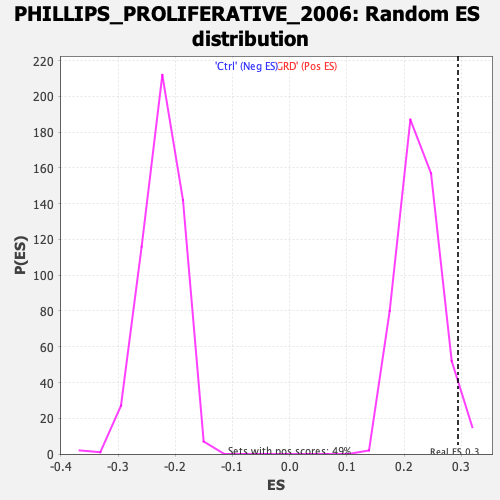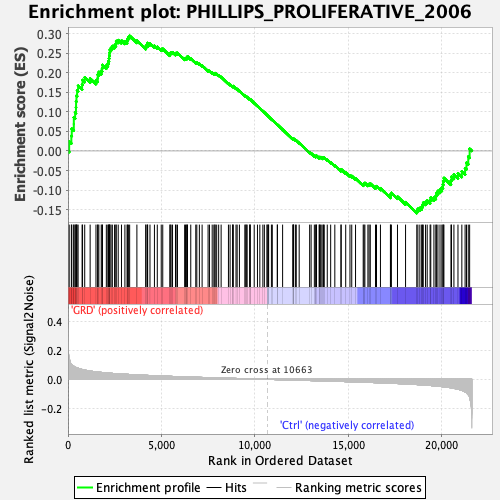
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | GRD |
| GeneSet | PHILLIPS_PROLIFERATIVE_2006 |
| Enrichment Score (ES) | 0.2944577 |
| Normalized Enrichment Score (NES) | 1.2969382 |
| Nominal p-value | 0.040567953 |
| FDR q-value | 0.10944437 |
| FWER p-Value | 0.657 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RRM2 | RRM2 | 63 | 0.143 | 0.0243 | Yes |
| 2 | CREBZF | CREBZF | 179 | 0.103 | 0.0386 | Yes |
| 3 | MAD2L1 | MAD2L1 | 202 | 0.100 | 0.0567 | Yes |
| 4 | WEE1 | WEE1 | 311 | 0.089 | 0.0686 | Yes |
| 5 | CDCA7 | CDCA7 | 312 | 0.089 | 0.0856 | Yes |
| 6 | PEG10 | PEG10 | 381 | 0.083 | 0.0984 | Yes |
| 7 | SMC2 | SMC2 | 426 | 0.081 | 0.1118 | Yes |
| 8 | PCNA | PCNA | 432 | 0.081 | 0.1270 | Yes |
| 9 | ABHD3 | ABHD3 | 452 | 0.080 | 0.1414 | Yes |
| 10 | MNS1 | MNS1 | 487 | 0.078 | 0.1547 | Yes |
| 11 | TMEM14B | TMEM14B | 540 | 0.076 | 0.1668 | Yes |
| 12 | PAIP1 | PAIP1 | 754 | 0.067 | 0.1697 | Yes |
| 13 | TOM1L1 | TOM1L1 | 778 | 0.067 | 0.1814 | Yes |
| 14 | C3orf14 | C3orf14 | 894 | 0.063 | 0.1881 | Yes |
| 15 | CENPK | CENPK | 1184 | 0.057 | 0.1854 | Yes |
| 16 | CDKN2C | CDKN2C | 1496 | 0.051 | 0.1807 | Yes |
| 17 | ACN9 | ACN9 | 1589 | 0.050 | 0.1859 | Yes |
| 18 | DBF4 | DBF4 | 1593 | 0.050 | 0.1953 | Yes |
| 19 | MCM2 | MCM2 | 1649 | 0.049 | 0.2021 | Yes |
| 20 | FBXO5 | FBXO5 | 1778 | 0.047 | 0.2052 | Yes |
| 21 | GGH | GGH | 1825 | 0.047 | 0.2119 | Yes |
| 22 | YEATS4 | YEATS4 | 1843 | 0.046 | 0.2200 | Yes |
| 23 | ANKRD32 | ANKRD32 | 2050 | 0.044 | 0.2188 | Yes |
| 24 | DONSON | DONSON | 2132 | 0.043 | 0.2233 | Yes |
| 25 | KIAA0101 | KIAA0101 | 2165 | 0.043 | 0.2300 | Yes |
| 26 | EZH2 | EZH2 | 2191 | 0.043 | 0.2369 | Yes |
| 27 | GINS1 | GINS1 | 2208 | 0.042 | 0.2443 | Yes |
| 28 | LRIG3 | LRIG3 | 2227 | 0.042 | 0.2514 | Yes |
| 29 | CHEK1 | CHEK1 | 2240 | 0.042 | 0.2589 | Yes |
| 30 | CENPL | CENPL | 2302 | 0.041 | 0.2639 | Yes |
| 31 | TOP2A | TOP2A | 2371 | 0.040 | 0.2684 | Yes |
| 32 | KNTC1 | KNTC1 | 2490 | 0.039 | 0.2704 | Yes |
| 33 | TCF19 | TCF19 | 2561 | 0.039 | 0.2746 | Yes |
| 34 | MND1 | MND1 | 2576 | 0.039 | 0.2813 | Yes |
| 35 | PPIG | PPIG | 2685 | 0.038 | 0.2835 | Yes |
| 36 | FANCI | FANCI | 2860 | 0.036 | 0.2823 | Yes |
| 37 | CEP152 | CEP152 | 3043 | 0.035 | 0.2805 | Yes |
| 38 | CDK2 | CDK2 | 3158 | 0.034 | 0.2817 | Yes |
| 39 | XRCC4 | XRCC4 | 3187 | 0.034 | 0.2868 | Yes |
| 40 | GINS2 | GINS2 | 3229 | 0.033 | 0.2913 | Yes |
| 41 | ITGA2 | ITGA2 | 3297 | 0.033 | 0.2945 | Yes |
| 42 | PLK4 | PLK4 | 3684 | 0.030 | 0.2822 | No |
| 43 | CCNA2 | CCNA2 | 4156 | 0.027 | 0.2654 | No |
| 44 | LMNB1 | LMNB1 | 4174 | 0.027 | 0.2698 | No |
| 45 | ZWILCH | ZWILCH | 4254 | 0.027 | 0.2712 | No |
| 46 | TEX9 | TEX9 | 4258 | 0.027 | 0.2762 | No |
| 47 | NPHP1 | NPHP1 | 4384 | 0.026 | 0.2753 | No |
| 48 | RFC4 | RFC4 | 4622 | 0.025 | 0.2689 | No |
| 49 | MELK | MELK | 4775 | 0.024 | 0.2663 | No |
| 50 | DSN1 | DSN1 | 4986 | 0.023 | 0.2608 | No |
| 51 | CDC6 | CDC6 | 5067 | 0.022 | 0.2613 | No |
| 52 | ATG12 | ATG12 | 5450 | 0.020 | 0.2474 | No |
| 53 | PABPC4L | PABPC4L | 5465 | 0.020 | 0.2506 | No |
| 54 | TRMT6 | TRMT6 | 5514 | 0.020 | 0.2522 | No |
| 55 | TTK | TTK | 5590 | 0.020 | 0.2524 | No |
| 56 | CENPA | CENPA | 5748 | 0.019 | 0.2487 | No |
| 57 | TTC12 | TTC12 | 5781 | 0.019 | 0.2507 | No |
| 58 | USP1 | USP1 | 5854 | 0.018 | 0.2509 | No |
| 59 | CDC25C | CDC25C | 6238 | 0.017 | 0.2362 | No |
| 60 | HMGB2 | HMGB2 | 6285 | 0.017 | 0.2372 | No |
| 61 | RRM1 | RRM1 | 6350 | 0.016 | 0.2373 | No |
| 62 | CCDC34 | CCDC34 | 6358 | 0.016 | 0.2401 | No |
| 63 | ORC6L | ORC6L | 6391 | 0.016 | 0.2417 | No |
| 64 | SHFM1 | SHFM1 | 6565 | 0.015 | 0.2365 | No |
| 65 | NASP | NASP | 6833 | 0.014 | 0.2267 | No |
| 66 | KIF4A | KIF4A | 6889 | 0.014 | 0.2268 | No |
| 67 | RAD51AP1 | RAD51AP1 | 7025 | 0.013 | 0.2231 | No |
| 68 | GJC1 | GJC1 | 7172 | 0.013 | 0.2187 | No |
| 69 | MLF1 | MLF1 | 7491 | 0.012 | 0.2060 | No |
| 70 | SLC25A24 | SLC25A24 | 7565 | 0.011 | 0.2048 | No |
| 71 | MLF1IP | MLF1IP | 7711 | 0.011 | 0.2000 | No |
| 72 | DHFR | DHFR | 7801 | 0.010 | 0.1979 | No |
| 73 | CENPI | CENPI | 7859 | 0.010 | 0.1972 | No |
| 74 | RBBP8 | RBBP8 | 7862 | 0.010 | 0.1990 | No |
| 75 | SMC4 | SMC4 | 7951 | 0.010 | 0.1968 | No |
| 76 | KIF23 | KIF23 | 8057 | 0.009 | 0.1937 | No |
| 77 | TTC26 | TTC26 | 8189 | 0.009 | 0.1893 | No |
| 78 | MAGOHB | MAGOHB | 8580 | 0.008 | 0.1725 | No |
| 79 | BARD1 | BARD1 | 8668 | 0.007 | 0.1698 | No |
| 80 | C3orf34 | C3orf34 | 8803 | 0.007 | 0.1648 | No |
| 81 | MDFIC | MDFIC | 8822 | 0.007 | 0.1652 | No |
| 82 | PPIL5 | PPIL5 | 8833 | 0.007 | 0.1660 | No |
| 83 | RPA3 | RPA3 | 8985 | 0.006 | 0.1601 | No |
| 84 | HELLS | HELLS | 9045 | 0.006 | 0.1584 | No |
| 85 | TMEM79 | TMEM79 | 9167 | 0.005 | 0.1538 | No |
| 86 | C4orf47 | C4orf47 | 9457 | 0.004 | 0.1412 | No |
| 87 | NUSAP1 | NUSAP1 | 9506 | 0.004 | 0.1397 | No |
| 88 | E2F1 | E2F1 | 9534 | 0.004 | 0.1392 | No |
| 89 | NCAPH | NCAPH | 9580 | 0.004 | 0.1378 | No |
| 90 | GCLM | GCLM | 9716 | 0.003 | 0.1321 | No |
| 91 | CHAF1B | CHAF1B | 9740 | 0.003 | 0.1317 | No |
| 92 | CACYBP | CACYBP | 9742 | 0.003 | 0.1322 | No |
| 93 | TRIM36 | TRIM36 | 9958 | 0.002 | 0.1227 | No |
| 94 | CHAF1A | CHAF1A | 10132 | 0.002 | 0.1150 | No |
| 95 | SGOL2 | SGOL2 | 10259 | 0.001 | 0.1093 | No |
| 96 | ABCA5 | ABCA5 | 10421 | 0.001 | 0.1020 | No |
| 97 | EMP2 | EMP2 | 10501 | 0.001 | 0.0984 | No |
| 98 | SPIN4 | SPIN4 | 10648 | 0.000 | 0.0916 | No |
| 99 | SIP1 | SIP1 | 10649 | 0.000 | 0.0916 | No |
| 100 | TYMS | TYMS | 10714 | -0.000 | 0.0886 | No |
| 101 | PIN4 | PIN4 | 10730 | -0.000 | 0.0880 | No |
| 102 | LSM5 | LSM5 | 10878 | -0.001 | 0.0812 | No |
| 103 | HMMR | HMMR | 10925 | -0.001 | 0.0792 | No |
| 104 | KIAA0895 | KIAA0895 | 11192 | -0.002 | 0.0672 | No |
| 105 | NCAPG | NCAPG | 11200 | -0.002 | 0.0672 | No |
| 106 | EVC2 | EVC2 | 11477 | -0.003 | 0.0549 | No |
| 107 | KIF14 | KIF14 | 12031 | -0.005 | 0.0299 | No |
| 108 | C17orf75 | C17orf75 | 12035 | -0.005 | 0.0307 | No |
| 109 | E2F8 | E2F8 | 12050 | -0.005 | 0.0310 | No |
| 110 | RBM24 | RBM24 | 12064 | -0.005 | 0.0313 | No |
| 111 | FANCD2 | FANCD2 | 12160 | -0.005 | 0.0278 | No |
| 112 | ZC3HAV1L | ZC3HAV1L | 12206 | -0.005 | 0.0268 | No |
| 113 | CDC2 | CDC2 | 12363 | -0.006 | 0.0206 | No |
| 114 | SHOX2 | SHOX2 | 12923 | -0.008 | -0.0040 | No |
| 115 | ANKRD5 | ANKRD5 | 13001 | -0.008 | -0.0060 | No |
| 116 | EFCAB2 | EFCAB2 | 13174 | -0.009 | -0.0123 | No |
| 117 | PDK1 | PDK1 | 13252 | -0.009 | -0.0142 | No |
| 118 | GMPS | GMPS | 13258 | -0.009 | -0.0126 | No |
| 119 | DLGAP5 | DLGAP5 | 13290 | -0.009 | -0.0123 | No |
| 120 | FBXO11 | FBXO11 | 13432 | -0.010 | -0.0170 | No |
| 121 | TMEM38B | TMEM38B | 13445 | -0.010 | -0.0157 | No |
| 122 | PAWR | PAWR | 13492 | -0.010 | -0.0159 | No |
| 123 | EXOSC9 | EXOSC9 | 13539 | -0.010 | -0.0161 | No |
| 124 | MAGOH | MAGOH | 13631 | -0.011 | -0.0183 | No |
| 125 | NUF2 | NUF2 | 13660 | -0.011 | -0.0176 | No |
| 126 | BRCA1 | BRCA1 | 13703 | -0.011 | -0.0175 | No |
| 127 | TIMELESS | TIMELESS | 13865 | -0.011 | -0.0228 | No |
| 128 | ERCC6L | ERCC6L | 14047 | -0.012 | -0.0289 | No |
| 129 | C1orf133 | C1orf133 | 14269 | -0.013 | -0.0367 | No |
| 130 | CKS2 | CKS2 | 14596 | -0.014 | -0.0492 | No |
| 131 | DTL | DTL | 14609 | -0.014 | -0.0470 | No |
| 132 | HAUS6 | HAUS6 | 14851 | -0.015 | -0.0554 | No |
| 133 | ZWINT | ZWINT | 15079 | -0.016 | -0.0628 | No |
| 134 | PRIM2 | PRIM2 | 15174 | -0.017 | -0.0640 | No |
| 135 | HJURP | HJURP | 15377 | -0.018 | -0.0701 | No |
| 136 | CDKN2A | CDKN2A | 15787 | -0.019 | -0.0854 | No |
| 137 | MTF2 | MTF2 | 15829 | -0.020 | -0.0836 | No |
| 138 | C3orf15 | C3orf15 | 15876 | -0.020 | -0.0819 | No |
| 139 | MCM6 | MCM6 | 16039 | -0.021 | -0.0856 | No |
| 140 | ECT2 | ECT2 | 16101 | -0.021 | -0.0844 | No |
| 141 | PPIC | PPIC | 16165 | -0.021 | -0.0833 | No |
| 142 | ZNF367 | ZNF367 | 16445 | -0.022 | -0.0921 | No |
| 143 | CENPN | CENPN | 16498 | -0.023 | -0.0901 | No |
| 144 | SPC24 | SPC24 | 16709 | -0.024 | -0.0954 | No |
| 145 | AURKA | AURKA | 17242 | -0.027 | -0.1152 | No |
| 146 | WDR34 | WDR34 | 17253 | -0.027 | -0.1105 | No |
| 147 | DEK | DEK | 17294 | -0.027 | -0.1073 | No |
| 148 | CENPF | CENPF | 17620 | -0.029 | -0.1170 | No |
| 149 | RECQL | RECQL | 18052 | -0.031 | -0.1311 | No |
| 150 | TMPO | TMPO | 18652 | -0.036 | -0.1523 | No |
| 151 | IL13RA2 | IL13RA2 | 18711 | -0.036 | -0.1481 | No |
| 152 | RAD51 | RAD51 | 18816 | -0.037 | -0.1459 | No |
| 153 | BUB1 | BUB1 | 18913 | -0.038 | -0.1432 | No |
| 154 | E2F7 | E2F7 | 18955 | -0.038 | -0.1378 | No |
| 155 | C6orf173 | C6orf173 | 18998 | -0.039 | -0.1324 | No |
| 156 | CENPE | CENPE | 19121 | -0.040 | -0.1304 | No |
| 157 | TIFA | TIFA | 19209 | -0.041 | -0.1268 | No |
| 158 | IFT74 | IFT74 | 19364 | -0.042 | -0.1259 | No |
| 159 | CCNB1 | CCNB1 | 19401 | -0.042 | -0.1195 | No |
| 160 | ASPM | ASPM | 19569 | -0.044 | -0.1189 | No |
| 161 | CCNE2 | CCNE2 | 19671 | -0.046 | -0.1149 | No |
| 162 | NDC80 | NDC80 | 19700 | -0.046 | -0.1075 | No |
| 163 | STIL | STIL | 19782 | -0.047 | -0.1023 | No |
| 164 | KIF18A | KIF18A | 19894 | -0.048 | -0.0982 | No |
| 165 | CCDC138 | CCDC138 | 19994 | -0.050 | -0.0933 | No |
| 166 | C1orf112 | C1orf112 | 20051 | -0.051 | -0.0863 | No |
| 167 | C12orf48 | C12orf48 | 20069 | -0.051 | -0.0774 | No |
| 168 | TMEM106C | TMEM106C | 20103 | -0.051 | -0.0691 | No |
| 169 | NEK2 | NEK2 | 20480 | -0.058 | -0.0757 | No |
| 170 | NUCB2 | NUCB2 | 20509 | -0.058 | -0.0658 | No |
| 171 | C1orf97 | C1orf97 | 20638 | -0.062 | -0.0601 | No |
| 172 | ARNTL2 | ARNTL2 | 20855 | -0.068 | -0.0572 | No |
| 173 | ACYP1 | ACYP1 | 21062 | -0.075 | -0.0525 | No |
| 174 | WDHD1 | WDHD1 | 21236 | -0.086 | -0.0441 | No |
| 175 | PRPS2 | PRPS2 | 21310 | -0.092 | -0.0301 | No |
| 176 | ITGB3BP | ITGB3BP | 21405 | -0.105 | -0.0144 | No |
| 177 | EIF1AX | EIF1AX | 21479 | -0.122 | 0.0054 | No |